| (2) AGO1.ip | (5) AGO2.ip | (8) B-CELL | (19) BRAIN | (2) CELL-LINE | (2) DCR.mut | (2) DGCR8.mut | (8) EMBRYO | (10) ESC | (3) FIBROBLAST | (1) HEART | (1) KIDNEY | (4) LIVER | (1) LYMPH | (15) OTHER | (5) OTHER.mut | (1) PIWI.ip | (1) PIWI.mut | (4) SKIN | (3) SPLEEN | (12) TESTES | (1) UTERUS |
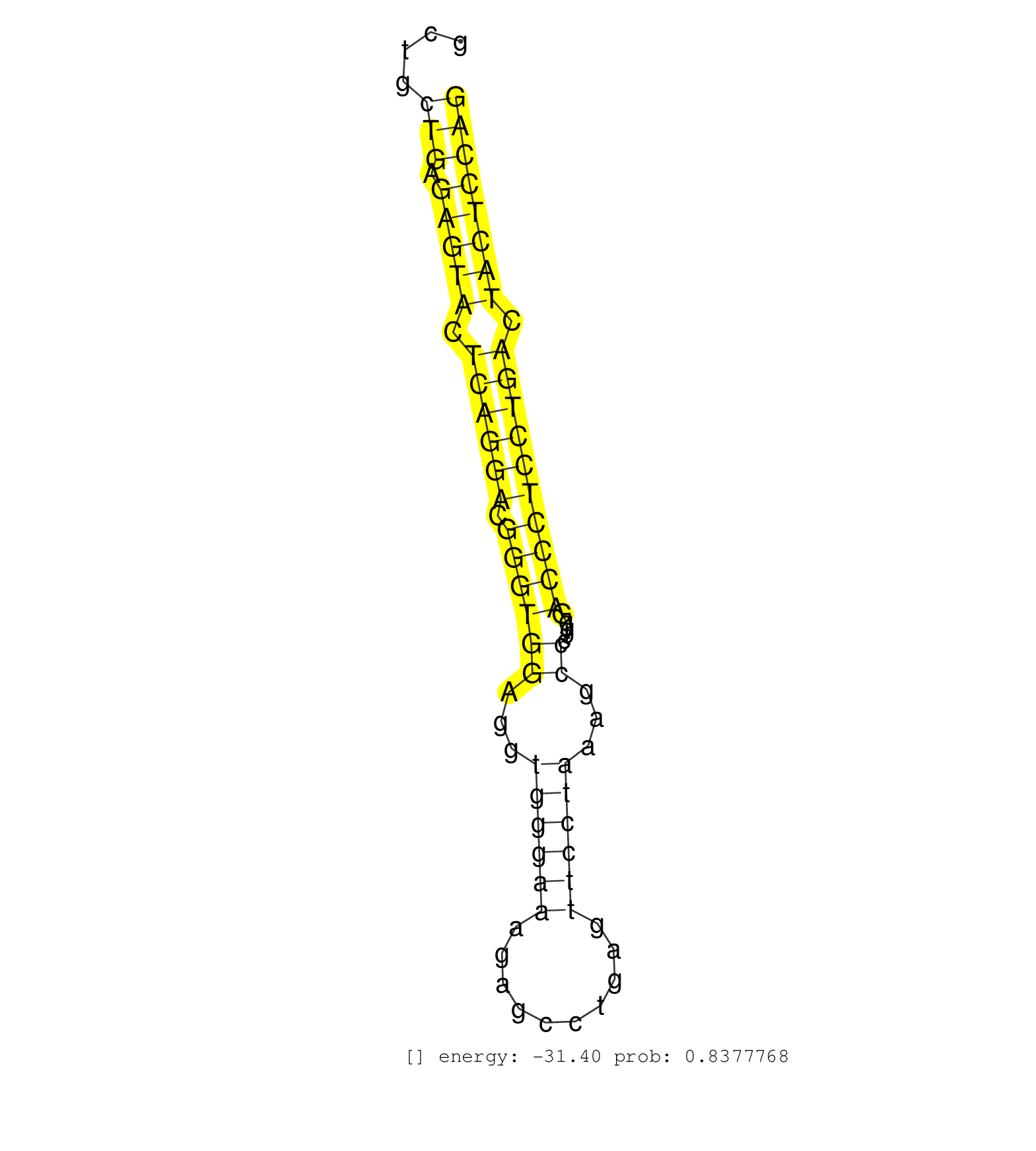
| GTGCCAGTGCCCATCCGTTACTGCATGCTGGGCAGCCGAGACTCCGCCAGGTCAGTTACATCATCTGCCCTGGTCCTGGCTTCCCTGGAATCCTTGGTTGGTTTGATCCAGCAAAGGCAGGGCCTGGAGTCCTTGGCTGCTGAGAGTACTCAGGACGGGTGGAGGTGGGAAAGAGCCTGAGTTCCTAAAGCCCTGGGACCCTCCTGACTACTCCAGGAACCCCCACACTCTGGTAGAAGTAACATCCTTTGCTGCCATCAACAAGT ...........................................................................................................................................(((.(((((.((((((.((((((...((((((..........))))))...)).....)))))))))).)))))))).................................................. .......................................................................................................................................136.............................................................................216................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR206939(GSM723280) other. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR206941(GSM723282) other. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM510444(GSM510444) brain_rep5. (brain) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | mjTestesWT3() Testes Data. (testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR065052(SRR065052) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | GSM361407(GSM361407) CGNP_P6_Ptc+-_Ink4c--_rep5. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | RuiDGCR8WT(Rui) DGCR8-dependent microRNA biogenesis is essent. (skin) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR073954(GSM629280) total RNA. (blood) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR525242(SRA056111/SRX170318) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR059769(GSM562831) Treg_Drosha. (spleen) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | GSM361415(GSM361415) WholeCerebellum_P6_p53--_Ink4c--_rep4. (brain) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTGGA....................................................................................................... | 23 | 1 | 69.00 | 69.00 | 7.00 | 1.00 | 2.00 | 5.00 | 2.00 | - | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | 2.00 | - | 2.00 | 1.00 | 1.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | - | 2.00 | 1.00 | - | 2.00 | 1.00 | - | - | 2.00 | - | 2.00 | 2.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGTGGA....................................................................................................... | 22 | 1 | 18.00 | 18.00 | 3.00 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GACCCTCCTGACTACTCCAGT................................................. | 21 | 1 | 8.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGTGG........................................................................................................ | 21 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTGG........................................................................................................ | 22 | 1 | 6.00 | 6.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GGAACCCCCACACTCTGGTA............................... | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TAAAGCCCTGGGACCCTCCTGACTACTCCAGA................................................. | 32 | 4.00 | 0.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGT.......................................................................................................... | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGTGGAT...................................................................................................... | 23 | 1 | 3.00 | 18.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGTGGAA...................................................................................................... | 23 | 1 | 3.00 | 18.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGTGGAAG..................................................................................................... | 24 | 1 | 3.00 | 18.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAGTACTCAGGACGGGTGGA....................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................ACACTCTGGTAGAAGTAACATC.................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GCCCATCCGTTACTGCATGCTG............................................................................................................................................................................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGCGGA....................................................................................................... | 23 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................CCACACTCTGGTAGAAGTAA........................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GTGGGAAAGAGCCTGGTT.................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................AGCCGAGACTCCGCCAG........................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTGGAT...................................................................................................... | 24 | 1 | 1.00 | 69.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................TACTGCATGCTGGGCAGCCG.................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GACCCTCCTGACTACTCCCGT................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................GAGAGTACTCAGGACGGGTG......................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................................................ACACTCTGGTAGAAGTAACATCCTTTGC.............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGCGG........................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................CTGGGCAGCCGAGACTCCTC........................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTT......................................................................................................... | 21 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGAGTCCTTGGCTGCTGA........................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTGGAA...................................................................................................... | 24 | 1 | 1.00 | 69.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TACTCAGGACGGGTGGAGGTGTGAA............................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................GCATGCTGGGCAGCCGAGACTCCG............................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGCTGGGCAGCCGAGACTCCGCCA......................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CATGCTGGGCAGCCGAGAC................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CAGGAACCCCCACACTCTGGTAGAAGT.......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGCGGA....................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTTGGCTGCTGAGAGTACT.................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGTGA........................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTTT........................................................................................................ | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GCTGGGCAGCCGAGACTCCGCCAGG....................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................AAGAGCCTGAGTTCCGGTT............................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................GCTGGGCAGCCGAGACTCCG............................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGAGTCCTTGGCTGCT............................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................ACCCTCCTGACTACTCCAGT................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGGGGA....................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTGAA....................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTGGAG...................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................TCTGGTAGAAGTAACATCCTTTGCTGCCATC....... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GACCCTCCTGACTACTCCAGA................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTGGCTGCTGAGAGAGT..................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGAGAGTACTCAGGACGGG........................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAGTACTCAGGACGGGTGGAG...................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGGGGG........................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GACCCTCCTGACTACTCCAG.................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTGGAGG..................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................GACCCTCCTGACTACTCCATTT................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................TACTGCATGCTGGGCAGCCGAGACTCCGC........................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAGAGTACTCAGGACGGG........................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................AACATCCTTTGCTGCCGGGC...... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTTGA....................................................................................................... | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TAAAGCCCTGGGACCCTCCTGACTACTCCAGT................................................. | 32 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................GAGAGTACTCAGGACGGGTGGAAT..................................................................................................... | 24 | 1 | 1.00 | 18.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................GGGCAGCCGAGACTCCGCC.......................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTGCCAGTGCCCATCCGTTACTGCATGCTGGGCAGCCGAGACTCCGCCAGGTCAGTTACATCATCTGCCCTGGTCCTGGCTTCCCTGGAATCCTTGGTTGGTTTGATCCAGCAAAGGCAGGGCCTGGAGTCCTTGGCTGCTGAGAGTACTCAGGACGGGTGGAGGTGGGAAAGAGCCTGAGTTCCTAAAGCCCTGGGACCCTCCTGACTACTCCAGGAACCCCCACACTCTGGTAGAAGTAACATCCTTTGCTGCCATCAACAAGT ...........................................................................................................................................(((.(((((.((((((.((((((...((((((..........))))))...)).....)))))))))).)))))))).................................................. .......................................................................................................................................136.............................................................................216................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR206939(GSM723280) other. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR206941(GSM723282) other. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM510444(GSM510444) brain_rep5. (brain) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | mjTestesWT3() Testes Data. (testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR065052(SRR065052) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | GSM361407(GSM361407) CGNP_P6_Ptc+-_Ink4c--_rep5. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | RuiDGCR8WT(Rui) DGCR8-dependent microRNA biogenesis is essent. (skin) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR073954(GSM629280) total RNA. (blood) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR525242(SRA056111/SRX170318) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR059769(GSM562831) Treg_Drosha. (spleen) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | GSM361415(GSM361415) WholeCerebellum_P6_p53--_Ink4c--_rep4. (brain) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................GAACCCCCACACTCTAAA................................ | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................CCCTGGAATCCTTGGGAA...................................................................................................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGAGAGTACTCAGGACGGGTGGA....................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................TAACATCCTTTGCTGAAA......... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TTACATCATCTGCCCAAGA................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................GGCAGCCGAGACTCCAC........................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................GTCCTGGCTTCCCTGGAAGGC............................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................CCTGAGTTCCTAAAGC........................................................................... | 16 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ..................................................................................................................................CCTTGGCTGCTGAGAGT....................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CCTGAGTTCCTAAAGCGCTC....................................................................... | 20 | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................CTGGTCCTGGCTTCCCT.................................................................................................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |