| (1) AGO.mut | (15) AGO2.ip | (29) BRAIN | (3) CELL-LINE | (1) DCR.mut | (2) DGCR8.mut | (10) EMBRYO | (7) ESC | (2) FIBROBLAST | (1) HEART | (1) KIDNEY | (5) LIVER | (1) LUNG | (9) OTHER | (4) OTHER.mut | (1) PIWI.mut | (3) SPLEEN | (11) TESTES | (1) THYMUS | (8) TOTAL-RNA | (1) UTERUS |
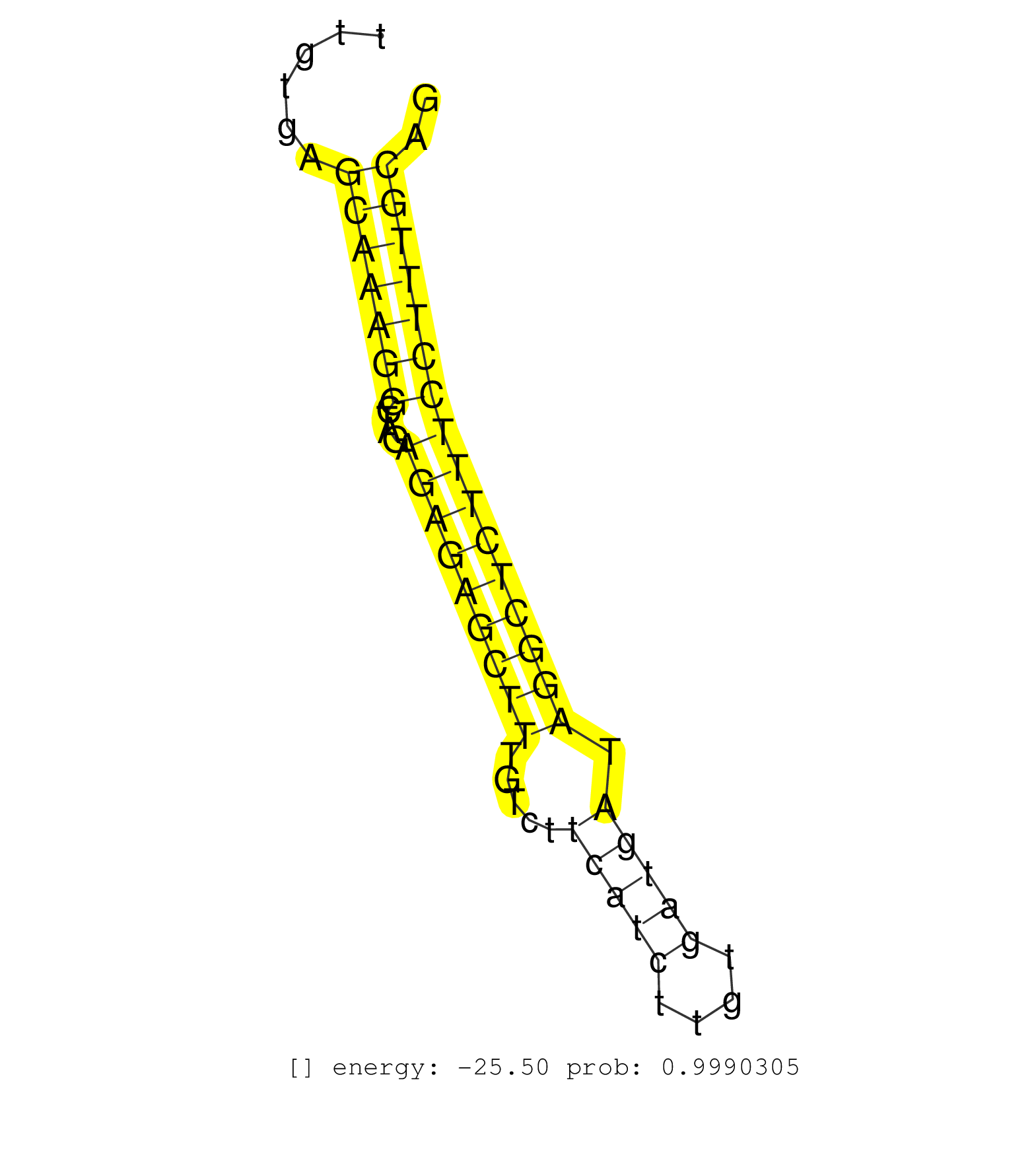
| GTTTATTTTTGCTGTTCTTGGTAAAAATACTGAAGTGCAGAAGATGGACATGTTCCAGAAAAGAAGATGGTAATTACAGGGTGGCATCTCTGTAACATAGTTATGTTCTGTACGTGTTGATGCTCCAGTTATTAAATTGTGAGCAAAGGCTAGAGAGAGCTTTGTCTTCATCTTGTGATGATAGGCTCTTTCCTTTGCAGCCAAACAGGCAAAAAGCTAATGGCGAAGTGTCGAATGCTTATCCAGGAGA ..............................................................................................................................................(((((((....(((((((((.....(((((....))))).)))))))))))))))).................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | mjTestesWT1() Testes Data. (testes) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | mjTestesWT4() Testes Data. (testes) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR206939(GSM723280) other. (brain) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | mjTestesWT3() Testes Data. (testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR206942(GSM723283) other. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR206940(GSM723281) other. (brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR206941(GSM723282) other. (brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306526(GSM750569) 19-24nt. (ago2 brain) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR037920(GSM510458) e12p5_rep2. (embryo) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR037909(GSM510446) newborn_rep2. (total RNA) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR073954(GSM629280) total RNA. (blood) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM510454(GSM510454) newborn_rep10. (total RNA) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR059765(GSM562827) DN3_control. (thymus) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR037902(GSM510438) testes_rep3. (testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGT................................................. | 21 | 1 | 80.00 | 27.00 | 16.00 | 9.00 | 7.00 | 9.00 | - | - | 2.00 | 1.00 | 8.00 | - | - | 3.00 | - | 1.00 | - | - | - | 3.00 | - | - | - | 2.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGA................................................. | 21 | 1 | 73.00 | 27.00 | 14.00 | 13.00 | 11.00 | 10.00 | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | 2.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 3.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TAGGCTCTTTCCTTTGCAGT................................................. | 20 | 1 | 31.00 | 3.00 | - | 1.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 4.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | 3.00 | - | - | - | 1.00 | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAG.................................................. | 20 | 1 | 27.00 | 27.00 | - | - | - | - | - | - | 3.00 | 1.00 | - | 6.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| .............................................................................................................................................AGCAAAGGCTAGAGAGAGCTTTGT..................................................................................... | 24 | 1 | 23.00 | 23.00 | - | - | - | - | 14.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GATAGGCTCTTTCCTTTGCAGA................................................. | 22 | 12.00 | 0.00 | - | 1.00 | 1.00 | - | - | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................AGCAAAGGCTAGAGAGAGCTTTGA..................................................................................... | 24 | 1 | 11.00 | 6.00 | - | - | - | - | - | - | - | 2.00 | - | - | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GATAGGCTCTTTCCTTTGCAGT................................................. | 22 | 8.00 | 0.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................AGCAAAGGCTAGAGAGAGCTTTG...................................................................................... | 23 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGATAGGCTCTTTCCTTTGCAG.................................................. | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................GTGATGATAGGCTCTTTCCTT....................................................... | 21 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TAGGCTCTTTCCTTTGCAG.................................................. | 19 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGTT................................................ | 22 | 1 | 3.00 | 27.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCACCT................................................ | 22 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................TAGGCTCTTTCCTTTGCAGA................................................. | 20 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGATAGGCTCTTTCCTTTGCAGA................................................. | 23 | 1 | 3.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TAGGCTCTTTCCTTTGCAGTT................................................ | 21 | 1 | 2.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGATAGGCTCTTTCCTTTGCAGT................................................. | 23 | 1 | 2.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................CAAAAAGCTAATGGCGAAG...................... | 19 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTTGATGCTCCAGTTATTAAATTGTGAGCAAAGGCTAGAGAGAGCTTTGTCTTCATCTTGTGATGATAGGCTCTTTCCTTTGCAG.................................................. | 86 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGTA................................................ | 22 | 1 | 2.00 | 27.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTCAGA.................................................. | 20 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................AAATTGTGAGCAAAGGCTAGAGA.............................................................................................. | 23 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TAGGCTCTTTCCTTTGCAGAAT............................................... | 22 | 1 | 2.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGATAGGCTCTTTCCTTTGCAGAA................................................ | 24 | 1 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGATAGGCTCTTTCCTTTGCAGAT................................................ | 24 | 1 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................GTGATGATAGGCTCTTTCCTTT...................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................AGATGGACATGTTCCAGAAAAGA.......................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GAGCAAAGGCTAGAGAGAGCTTT....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................AATTACAGGGTGGCATCTCTGT............................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................AGAGAGAGCTTTGTCTTCATCT............................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGCAAAGGCTAGAGAGAGCTTAGT..................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................CAGGCAAAAAGCTAATGGCGA........................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ACGTGTTGATGCTCCAGTTATT..................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AAGGCTAGAGAGAGCTTTG...................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGAT................................................ | 22 | 1 | 1.00 | 27.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ACGTGTTGATGCTCCAGTT........................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CAGGCAAAAAGCTAATGGCGAAGT..................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CAGGCAAAAAGCTAATGGCGAAGTG.................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GCTAATGGCGAAGTGTCGAA............... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGTAA............................................... | 23 | 1 | 1.00 | 27.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................AATACTGAAGTGCAGAAGAT............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGAA................................................ | 22 | 1 | 1.00 | 27.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGAAT............................................... | 23 | 1 | 1.00 | 27.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AAGCTAATGGCGAAGTGTC.................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TAGGCTCTTTCCTTTGCAGTGA............................................... | 22 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGCAAAGGCTAGAGAGAGCTTAA...................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGAG................................................ | 22 | 1 | 1.00 | 27.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GAGCAAAGGCTAGAGAGAGCTT........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGCAAAGGCTAGAGAGAGCTTTTA..................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TGATAGGCTCTTTCCTTTGCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAGCT................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................GATAGGCTCTTTCCTTTGCAGATT............................................... | 24 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................AGCAAAGGCTAGAGAGAG........................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATGTTCTGTACGTGTTGATGCTCCAGTTATTAAATTGTGAGCAAAGGCTAGAGAGAGCTTTGTCTTC................................................................................. | 67 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................AATGGCGAAGTGTCGAATGCTT.......... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CTAGAGAGAGCTTTGTCTT.................................................................................. | 19 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATGTTCTGTACGTGTTGATGCTCCA........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGTGATGATAGGCTCTTTCCTTTGCAGA................................................. | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ..........................................................................................................................................................................................................................AATGGCGAAGTGTCGAAT.............. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAGCAAAGGCTAGAGAGAGCTTTG...................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATAGGCTCTTTCCTTTGCAAT................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................TAGGCTCTTTCCTTTGCAGTA................................................ | 21 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................AGGGTGGCATCTCTGTAACATAG...................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGCAAAGGCTAGAGAGAGCTTTGAA.................................................................................... | 25 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................AATTACAGGGTGGCATCTCTGTAA........................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................ATTAAATTGTGAGCAAAGGCT................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CGAAGTGTCGAATGCTTA......... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ...............................................................................................................................................................................................................................CGAAGTGTCGAATGCTTATCCAGGAG. | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ...........................................................................................................................................................................................................................ATGGCGAAGTGTCGAATGC............ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................CGAATGCTTATCCAGGAGA | 19 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTTTATTTTTGCTGTTCTTGGTAAAAATACTGAAGTGCAGAAGATGGACATGTTCCAGAAAAGAAGATGGTAATTACAGGGTGGCATCTCTGTAACATAGTTATGTTCTGTACGTGTTGATGCTCCAGTTATTAAATTGTGAGCAAAGGCTAGAGAGAGCTTTGTCTTCATCTTGTGATGATAGGCTCTTTCCTTTGCAGCCAAACAGGCAAAAAGCTAATGGCGAAGTGTCGAATGCTTATCCAGGAGA ..............................................................................................................................................(((((((....(((((((((.....(((((....))))).)))))))))))))))).................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | mjTestesWT1() Testes Data. (testes) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | mjTestesWT4() Testes Data. (testes) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR206939(GSM723280) other. (brain) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | mjTestesWT3() Testes Data. (testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR206942(GSM723283) other. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR206940(GSM723281) other. (brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR206941(GSM723282) other. (brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306526(GSM750569) 19-24nt. (ago2 brain) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR037920(GSM510458) e12p5_rep2. (embryo) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR037909(GSM510446) newborn_rep2. (total RNA) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR073954(GSM629280) total RNA. (blood) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM510454(GSM510454) newborn_rep10. (total RNA) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR059765(GSM562827) DN3_control. (thymus) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR037902(GSM510438) testes_rep3. (testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................CTCCAGTTATTAAATGAGA............................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................AGGCTCTTTCCTTTGCAC.................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |