| (7) AGO2.ip | (2) AGO3.ip | (7) B-CELL | (10) BRAIN | (2) CELL-LINE | (1) DGCR8.mut | (3) EMBRYO | (7) ESC | (3) FIBROBLAST | (2) KIDNEY | (5) LIVER | (1) LUNG | (2) LYMPH | (19) OTHER | (4) OTHER.mut | (1) OVARY | (1) PIWI.ip | (2) SKIN | (5) SPLEEN | (11) TESTES |
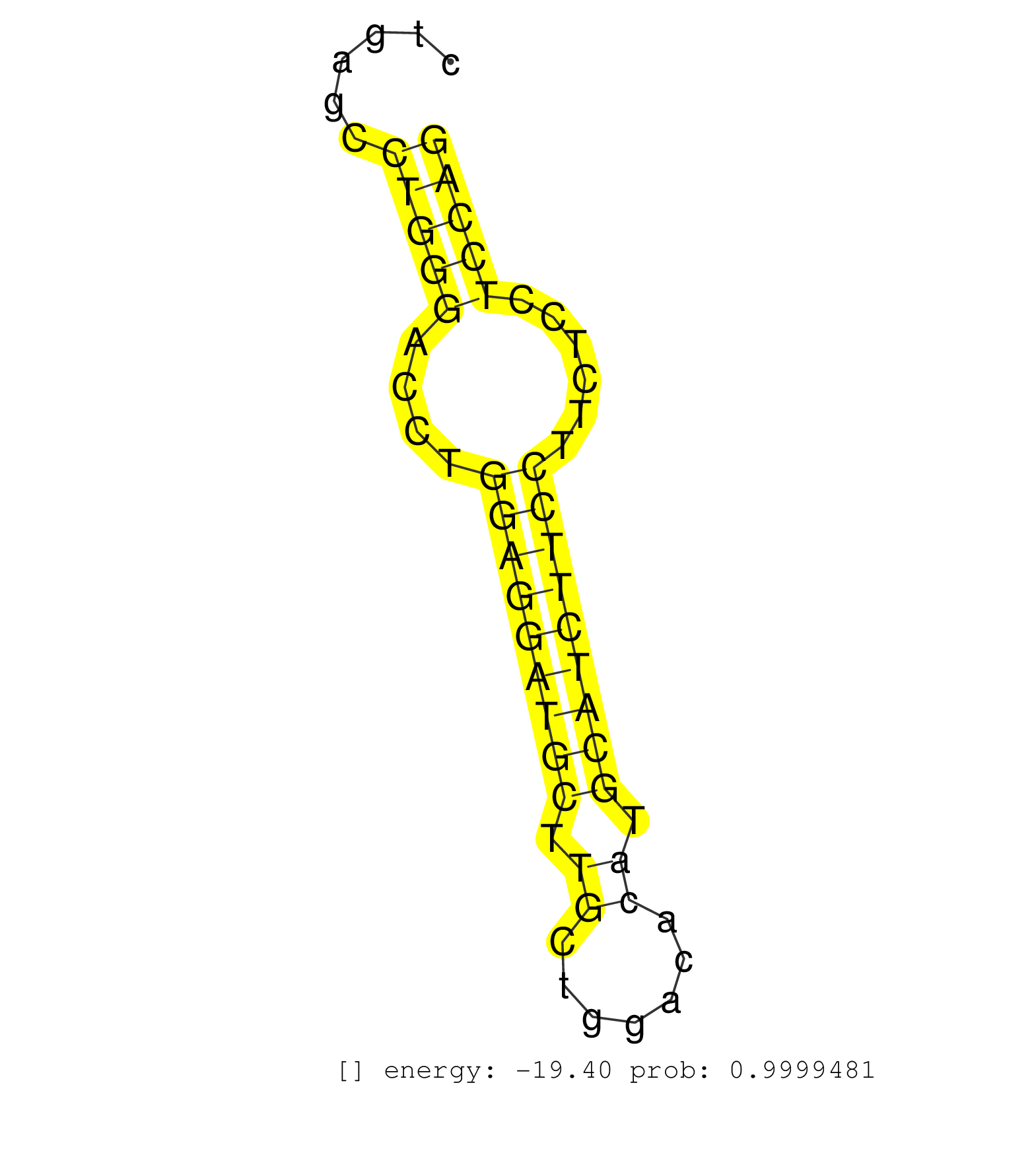
| TCCTCTGTGCTACGCACGGACAGATGAGGGGATTGCTGGGGTCTGGGTCTGCAGTGAGCAGAGCCAACAGGCTTCAGCAAACTGGTGCCCCCACTGCCAGGGCCCTGCTCCTTCCCTGTGGGCTCAGATGCTGCCTAGGTGCTCTGAGCCTGGGACCTGGAGGATGCTTGCTGGACACATGCATCTTCCTTCTCCTCCAGGAGCTGTATGCAGCTGGGGAGAACCGCCTGGGAACAGACGAGTCCAAGTT .....................................................................................................................................................(((((....(((((((((.((.......)).)))))))))......))))).................................................. ...............................................................................................................................................144.....................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | mjTestesWT4() Testes Data. (testes) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | mjLiverKO3() Liver Data. (Zcchc11 liver) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | mjTestesWT2() Testes Data. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | mjTestesWT3() Testes Data. (testes) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR065052(SRR065052) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR059774(GSM562836) MEF_control. (MEF) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | mjLiverWT3() Liver Data. (liver) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGT............................................................................... | 23 | 3 | 27.00 | 20.33 | 10.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTG................................................................................ | 22 | 3 | 20.33 | 20.33 | 1.33 | 1.00 | 1.33 | 0.67 | - | - | 1.33 | 3.33 | 0.33 | 1.00 | 0.33 | 0.67 | - | - | - | - | - | - | - | 0.67 | - | - | 0.67 | 0.67 | - | - | 0.33 | - | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | 0.67 | 0.67 | - | - | - | - | - | 0.33 | - | 0.33 | - | - | - | - | 0.33 | - | 0.33 | 0.33 | - | 0.33 | 0.33 | 0.33 | - | - | - | 0.33 | - | 0.33 | 0.33 | 0.33 | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTT................................................................................. | 21 | 3 | 8.67 | 8.67 | 1.33 | 1.33 | - | - | - | - | 0.33 | - | - | - | 0.33 | 0.67 | - | - | - | - | - | - | - | - | 0.33 | 0.67 | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | 0.33 | 0.33 | - | - | - | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | 0.33 |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGA............................................................................... | 23 | 3 | 8.00 | 20.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGCATCTTCCTTCTCCTCCAGT................................................. | 22 | 8.00 | 0.00 | - | - | - | - | 4.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TGCATCTTCCTTCTCCTCCAGAC................................................ | 23 | 4.00 | 0.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TGCATCTTCCTTCTCCTCCAGA................................................. | 22 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGTAT............................................................................. | 25 | 3 | 2.00 | 20.33 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTA................................................................................ | 22 | 3 | 2.00 | 8.67 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATGCATCTTCCTTCTCCTCCAGT................................................. | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTT................................................................................ | 22 | 3 | 2.00 | 8.67 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGTA.............................................................................. | 24 | 3 | 2.00 | 20.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGAA.............................................................................. | 24 | 3 | 2.00 | 20.33 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCT.................................................................................. | 20 | 3 | 2.00 | 2.00 | 0.33 | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CAGATGCTGCCTAGGTGCTCTGAGCCTGGGACCTGGAGGATGCTTGCTGGACACATGCATCTTCCTTCTCCTCCAG.................................................. | 76 | 3 | 1.67 | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GGAGAACCGCCTGGGAACAGACGA......... | 24 | 3 | 1.67 | 1.67 | - | - | - | - | - | - | - | - | - | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CTGGGACCTGGAGGATGCTTGT............................................................................... | 22 | 3 | 1.00 | 0.67 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTAG............................................................................... | 23 | 3 | 1.00 | 8.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGTT.............................................................................. | 24 | 3 | 1.00 | 20.33 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GCCTGGGACCTGGAGGATGCTTGA............................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGATTG................................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGTTAA................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTAAA............................................................................... | 23 | 3 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATGCATCTTCCTTCTCCTCCAGTTT............................................... | 25 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................TGGGACCTGGAGGATGCTTGCTT............................................................................. | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTAAT.............................................................................. | 24 | 3 | 1.00 | 8.67 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCAAAA............................................................................... | 23 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................ATGCATCTTCCTTCTCCTCCAGTT................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................GACAGATGAGGGGATTGTGG.................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCGTG................................................................................ | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................TCTGGGTCTGCAGTGAGTACG............................................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................AGCCTGGGACCTGGAGGATGCTTGTT.............................................................................. | 26 | 3 | 1.00 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GAGCCTGGGACCTGGCTA....................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTTT............................................................................... | 23 | 3 | 1.00 | 8.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTTAA.............................................................................. | 24 | 3 | 1.00 | 8.67 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGAT.............................................................................. | 24 | 3 | 1.00 | 20.33 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GCCTGGGACCTGGAGGATGCTTAT............................................................................... | 24 | 3 | 1.00 | 0.67 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGTAA............................................................................. | 25 | 3 | 1.00 | 20.33 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................GGGACCTGGAGGATGTGGC................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTTA............................................................................... | 23 | 3 | 1.00 | 8.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATGCATCTTCCTTCTCCTCTTT.................................................. | 22 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................CAGCTGGGGAGAACCGCC...................... | 18 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGCTACGCACGGACAGATGAGGGGA.......................................................................................................................................................................................................................... | 25 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CTGGGACCTGGAGGATGCTTG................................................................................ | 21 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - |
| ...................................................................................................................................................GCCTGGGACCTGGAGGATGCTT................................................................................. | 22 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GGACCTGGAGGATGCTTGCTGGACAC........................................................................ | 26 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AACCGCCTGGGAACAGACGAGT....... | 22 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................CGCCTGGGAACAGACGAG........ | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGAGCTGTATGCAGC.................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................CTGGAGGATGCTTGCTGGACACA....................................................................... | 23 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGTATGCAGCTGGGGAGAACC......................... | 21 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................ATGCAGCTGGGGAGAACCGCCTGGGAACAG............. | 30 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................GGGACCTGGAGGATGCTT................................................................................. | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GGAGAACCGCCTGGGAACAGACGAG........ | 25 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............ACGGACAGATGAGGGGATTGCTGG................................................................................................................................................................................................................... | 24 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - |
| ....................................................................................................................................................CCTGGGACCTGGAGGATGCTTGCTGG............................................................................ | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GGGAGAACCGCCTGGGAACAGACGA......... | 25 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AGCCTGGGACCTGGAGGATGCTTG................................................................................ | 24 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GGAGAACCGCCTGGGAACAGAC........... | 22 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCTGAGCCTGGGACCTGGAGGAT..................................................................................... | 23 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CTGTATGCAGCTGGGG............................... | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCTCTGTGCTACGCACGGACAGATGAGGGGATTGCTGGGGTCTGGGTCTGCAGTGAGCAGAGCCAACAGGCTTCAGCAAACTGGTGCCCCCACTGCCAGGGCCCTGCTCCTTCCCTGTGGGCTCAGATGCTGCCTAGGTGCTCTGAGCCTGGGACCTGGAGGATGCTTGCTGGACACATGCATCTTCCTTCTCCTCCAGGAGCTGTATGCAGCTGGGGAGAACCGCCTGGGAACAGACGAGTCCAAGTT .....................................................................................................................................................(((((....(((((((((.((.......)).)))))))))......))))).................................................. ...............................................................................................................................................144.....................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | mjTestesWT4() Testes Data. (testes) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | mjLiverKO3() Liver Data. (Zcchc11 liver) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | mjTestesWT2() Testes Data. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | mjTestesWT3() Testes Data. (testes) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR065052(SRR065052) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR059774(GSM562836) MEF_control. (MEF) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | mjLiverWT3() Liver Data. (liver) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................TGCTCCTTCCCTGTGGTAGA............................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................CCCCCACTGCCAGGGCGGA................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................CTTCCCTGTGGGCTCAGATGCTGCCTA................................................................................................................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - |