| (2) AGO1.ip | (10) AGO2.ip | (2) B-CELL | (18) BRAIN | (3) CELL-LINE | (1) DCR.mut | (1) DGCR8.mut | (10) EMBRYO | (7) ESC | (3) FIBROBLAST | (2) LIVER | (1) LYMPH | (6) OTHER | (3) OTHER.mut | (2) PIWI.ip | (2) SKIN | (3) SPLEEN | (11) TESTES |
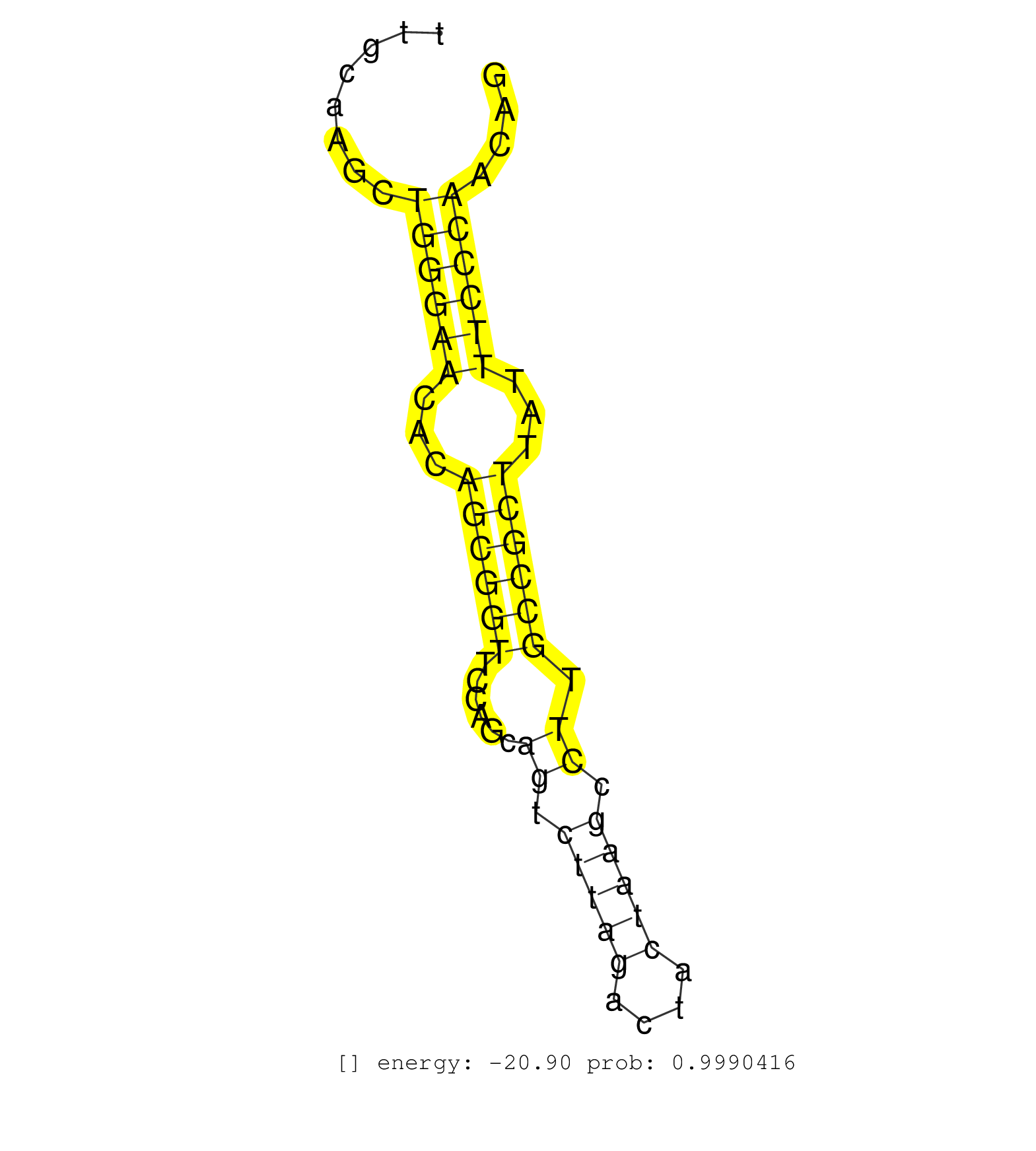
| TCTGACAAATGGACAGACAGACTTAGGAGAAATGCCTTAGTCTGCCAGCAGTCATTCGACTGCTACTGATGAAGATAGTCCCCACAGGCATCTGGCCACACTTTGGGCAGAGTGGGTGGCCCTGGGGATCTTTGCAAGCTGGGAACACAGCGGTTCCAGCAGTCTTAGACTACTAAGCCTTGCCGCTTATTTCCCAACAGTGGCGAGAAGAGCGCTTCAGACAAACCAGCGAAAGCACCAACCAGCGAGT ...........................................................................................................................................((((((...((((((......((.(((((....))))).)).))))))...))))))...................................................... ...................................................................................................................................132.................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | mjTestesWT4() Testes Data. (testes) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR059775(GSM562837) MEF_Drosha. (MEF) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR394083(GSM855969) "background strain: C57BL6/SV129cell type: KR. (cell line) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR206939(GSM723280) other. (brain) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR059769(GSM562831) Treg_Drosha. (spleen) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | RuiDGCR8KO(Rui) DGCR8-dependent microRNA biogenesis is essent. (dgcr8 skin) | SRR059770(GSM562832) Treg_Dicer. (spleen) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR206940(GSM723281) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | GSM475281(GSM475281) total RNA. (testes) | SRR553586(SRX182792) source: Testis. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR037930(GSM510468) e7p5_rep4. (embryo) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAACAGT................................................. | 23 | 1 | 38.00 | 38.00 | 1.00 | 5.00 | 2.00 | 3.00 | - | - | - | 3.00 | 2.00 | - | - | 1.00 | - | 2.00 | - | 2.00 | - | - | 1.00 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 2.00 | - | - | - | 2.00 | 1.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAACAGA................................................. | 23 | 1 | 16.00 | 5.00 | 6.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCCAG........................................................................................... | 23 | 1 | 8.00 | 8.00 | 1.00 | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCCAT........................................................................................... | 23 | 1 | 6.00 | 5.00 | - | - | - | - | - | - | - | - | - | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................GGGTGGCCCTGGGGATCTTTGCAAGC............................................................................................................... | 26 | 1 | 6.00 | 6.00 | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGCCGCTTATTTCCCAACAGA................................................. | 21 | 6.00 | 0.00 | - | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCCA............................................................................................ | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAACA................................................... | 21 | 1 | 5.00 | 5.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAACAG.................................................. | 22 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CTGGGAACACAGCGGTTCCAG........................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CTGCTACTGATGAAGGCT............................................................................................................................................................................. | 18 | 3.00 | 0.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................CTGGGAACACAGCGGTTCCAGCA......................................................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCC............................................................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AAGCTGGGAACACAGCGGTTCCAGC.......................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAACT................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAAC.................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCCAGA.......................................................................................... | 24 | 1 | 2.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTGCCGCTTATTTCCCAACAGA................................................. | 22 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..TGACAAATGGACAGACAGACTTAGGA.............................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CCGCTTATTTCCCAACAG.................................................. | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CTGCTACTGATGAAGGCTT............................................................................................................................................................................ | 19 | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................CTTCAGACAAACCAGCGAAA................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCTTGCCGCTTATTTCCCAACAGT................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................GGTTCCAGCAGTCTTAGACTA.............................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................ACAGACTTAGGAGAACCC........................................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................CGAGAAGAGCGCTTCAGACAAACCAGCGAAAGCACCA.......... | 37 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCTTGCCGCTTATTTCCCAACAG.................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AAGCTGGGAACACAGCGGTTCC............................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............GACAGACTTAGGAGAAAAGCT...................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAACCGT................................................. | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................CGAGAAGAGCGCTTCAGACAAACCAGC.................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AGCTGGGAACACAGCGGTT............................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................ACAGACTTAGGAGAACAC........................................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................CTGGGAACACAGCGGTTCCAGC.......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCCAGT.......................................................................................... | 24 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCAAG........................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAACGGT................................................. | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTGCCGCTTATTTCCCAACAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................CTTCAGACAAACCAGCGAAAGCACTAAC........ | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCCAGTA......................................................................................... | 25 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCTTGCCGCTTATTTCCCAAC.................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................ATCTTTGCAAGCTGGGAACACAGCGGTTCCAGCAGTCTTAGACTACTAAGCCTTGCCGCTTATTTCCCAACA................................................... | 72 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................CTGGGAACACAGCGGTTCCCGCA......................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................CCTTGCCGCTTATTTCCCAACA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AGCTGGGAACACAGCGGTTCCAA........................................................................................... | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTTGCCGCTTATTTCCCAACAT.................................................. | 22 | 1 | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................GAGAAGAGCGCTTCAGAC............................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......AAATGGACAGACAGACTT.................................................................................................................................................................................................................................. | 18 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTGACAAATGGACAGACAGACTTAGGAGAAATGCCTTAGTCTGCCAGCAGTCATTCGACTGCTACTGATGAAGATAGTCCCCACAGGCATCTGGCCACACTTTGGGCAGAGTGGGTGGCCCTGGGGATCTTTGCAAGCTGGGAACACAGCGGTTCCAGCAGTCTTAGACTACTAAGCCTTGCCGCTTATTTCCCAACAGTGGCGAGAAGAGCGCTTCAGACAAACCAGCGAAAGCACCAACCAGCGAGT ...........................................................................................................................................((((((...((((((......((.(((((....))))).)).))))))...))))))...................................................... ...................................................................................................................................132.................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | mjTestesWT4() Testes Data. (testes) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR059775(GSM562837) MEF_Drosha. (MEF) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR394083(GSM855969) "background strain: C57BL6/SV129cell type: KR. (cell line) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR206939(GSM723280) other. (brain) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR059769(GSM562831) Treg_Drosha. (spleen) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | RuiDGCR8KO(Rui) DGCR8-dependent microRNA biogenesis is essent. (dgcr8 skin) | SRR059770(GSM562832) Treg_Dicer. (spleen) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR206940(GSM723281) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | GSM475281(GSM475281) total RNA. (testes) | SRR553586(SRX182792) source: Testis. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR037930(GSM510468) e7p5_rep4. (embryo) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................CTGGGGATCTTTGCATCCT.............................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................GTCTGCCAGCAGTCAGTG................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................CTGGGGATCTTTGCACTGT.............................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTATTTCCCAACAGTGAT.............................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ...........................................................CTGCTACTGATGAAGGCC............................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................CTTCAGACAAACCAGCGAAAGCACCA.......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |