| (1) AGO.mut | (2) AGO1.ip | (15) AGO2.ip | (5) B-CELL | (31) BRAIN | (3) CELL-LINE | (1) DCR.mut | (2) DGCR8.mut | (6) EMBRYO | (6) ESC | (4) FIBROBLAST | (1) KIDNEY | (8) LIVER | (3) LYMPH | (10) OTHER | (2) OTHER.mut | (2) PIWI.ip | (1) PIWI.mut | (3) SKIN | (2) SPLEEN | (11) TESTES | (1) TOTAL-RNA | (2) UTERUS |
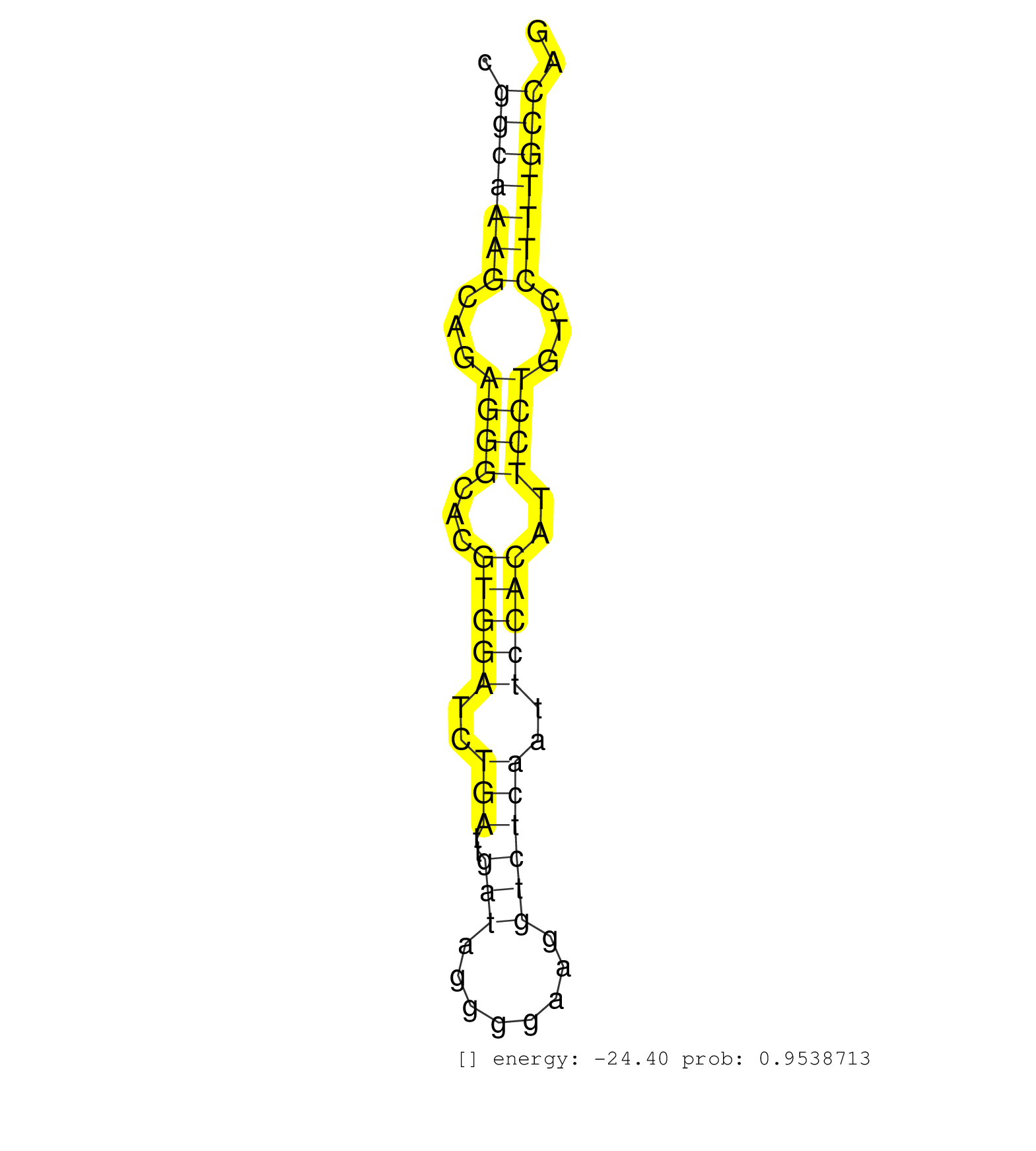
| CCGGGAGTGAGCGGGCTGACTCCTCAGGGGCCCGGGGGATGCGTCTAAAGGTGAGGATGGTGGATGCCTCAGGGGCTGTGCAGGGAGGGAGGTTGTTGATCGTCCCCTAGCACCCTTGCCGGCAAAGCAGAGGGCACGTGGATCTGATTGATAGGGGAAGGTCTCAATTCCACATTCCTGTCCTTTGCCAGGAAGGCGCCAACATCAATAAGTCCCTGACTACGCTTGGGAAGGTGATCTC ........................................................................................................................(((((((...((((...(((((..(((..(((........))))))..)))))..))))...))))))).................................................... .......................................................................................................................120....................................................................191................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR394084(GSM855970) "background strain: C57BL6/SV129cell type: KR. (dicer cell line) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR206940(GSM723281) other. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM509280(GSM509280) small RNA cloning by length. (testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR065049(SRR065049) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR037912(GSM510449) newborn_rep5. (total RNA) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037920(GSM510458) e12p5_rep2. (embryo) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR206941(GSM723282) other. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR037902(GSM510438) testes_rep3. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................CACATTCCTGTCCTTTGCCAGT................................................. | 22 | 1 | 40.00 | 1.00 | 4.00 | - | 7.00 | - | - | - | - | 3.00 | - | 1.00 | - | - | - | 1.00 | 3.00 | - | - | - | 3.00 | - | - | 2.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGT.............................................................................................. | 23 | 1 | 8.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCT................................................................................................ | 21 | 1 | 8.00 | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AGCAGAGGGCACGTGGATCTG............................................................................................... | 21 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................................ATTCCACATTCCTGTCCTTTGC..................................................... | 22 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................ACATTCCTGTCCTTTGCCAGT................................................. | 21 | 6.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .............................................................................................................................AGCAGAGGGCACGTGGATCTGT.............................................................................................. | 22 | 1 | 6.00 | 7.00 | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AGCAGAGGGCACGTGGATCTGA.............................................................................................. | 22 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................ATTCCACATTCCTGTCCTTTGCCAGT................................................. | 26 | 1 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGA.............................................................................................. | 23 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................GAGGGCACGTGGATCTGAA............................................................................................. | 19 | 4.00 | 0.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................CAGGGGCCCGGGGGATGCGTCTAAAGG.............................................................................................................................................................................................. | 27 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................ATTCCACATTCCTGTCCTTTGCC.................................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGACTACGCTTGGGAAG........ | 17 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................CCTGACTACGCTTGGGAA......... | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................ATTCCACATTCCTGTCCTTTGCCA................................................... | 24 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CGGCAAAGCAGAGGGCACGTGGAA.................................................................................................. | 24 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................AGCAGAGGGCACGTGGATCT................................................................................................ | 20 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGTT............................................................................................. | 24 | 1 | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AGCAGAGGGCACGTGGATCTGTAA............................................................................................ | 24 | 1 | 2.00 | 7.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CACATTCCTGTCCTTTGCCAGA................................................. | 22 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AGCAGAGGGCACGTGGATCTGTA............................................................................................. | 23 | 1 | 2.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTT............................................................................................... | 22 | 1 | 2.00 | 8.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGAT............................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGAAA............................................................................................ | 25 | 1 | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGAGGGAGGTTGTTGGAT............................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................CCGGCAAAGCAGAGGGCACGTGG.................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................ATTCCACATTCCTGTCCTTTG...................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATAT................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................CAGGGGCCCGGGGGATTCGT..................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................AGCAGAGGGCACGTGGATC................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GACTCCTCAGGGGCCCGGGGGAT......................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CTCAGGGGCCCGGGGGATGCGTCTAAAGGAAG........................................................................................................................................................................................... | 32 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........GAGCGGGCTGACTCCTCAGGGGCCCGGGGG........................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGTAAA........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................GCAGAGGGCACGTGGCGCA................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................AGTCCCTGACTACGCTTGG............ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................GCCCGGGGGATGCGTCTAAAGGA............................................................................................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................GCCGGCAAAGCAGAGGGCACGT...................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................GCAGAGGGCACGTGGGGCA................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGAG............................................................................................. | 24 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TCAGGGGCTGTGCAGGGAGGGAGGT.................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GAGTGAGCGGGCTGACTCCTCAGGG.................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CTACGCTTGGGAAGGTGATC.. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AGCAGAGGGCACGTGGATCTGATTGA.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......GTGAGCGGGCTGACTCCTCAGG..................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GAGTGAGCGGGCTGACTCCTCAGGGGC.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AGCAGAGGGCACGTGGATCTTTAT............................................................................................ | 24 | 1 | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTAT.............................................................................................. | 23 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CACATTCCTGTCCTTTGCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................GGCAAAGCAGAGGGCACGTGA.................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................GCCGGCAAAGCAGAGGGCACGTGG.................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGAGCTT............................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................TGAGGATGGTGGATGATG............................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................AGGGCACGTGGATCTGT.............................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGTA............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CGGCAAAGCAGAGGGCACGTGGAG.................................................................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................CCGGCAAAGCAGAGGGCACGTGGATC................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....GAGTGAGCGGGCTGACTCCTCAGG..................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGACTACGCTTGGGAAGGTGATCTC | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTG............................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CCCTGACTACGCTTGGGAAGG....... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AAGCAGAGGGCACGTGGATCTGAA............................................................................................. | 24 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TAAGTCCCTGACTACGCTTGGGAAGG....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................GCAGAGGGCACGTGGAGCC................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................ATTCCACATTCCTGTCCTTTGCCAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGAGGGAGGTTGTTGTG............................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .CGGGAGTGAGCGGGCT................................................................................................................................................................................................................................ | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GGGGCCCGGGGGAT......................................................................................................................................................................................................... | 14 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CAGGGGCTGTGCAGGG............................................................................................................................................................ | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCGGGAGTGAGCGGGCTGACTCCTCAGGGGCCCGGGGGATGCGTCTAAAGGTGAGGATGGTGGATGCCTCAGGGGCTGTGCAGGGAGGGAGGTTGTTGATCGTCCCCTAGCACCCTTGCCGGCAAAGCAGAGGGCACGTGGATCTGATTGATAGGGGAAGGTCTCAATTCCACATTCCTGTCCTTTGCCAGGAAGGCGCCAACATCAATAAGTCCCTGACTACGCTTGGGAAGGTGATCTC ........................................................................................................................(((((((...((((...(((((..(((..(((........))))))..)))))..))))...))))))).................................................... .......................................................................................................................120....................................................................191................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR394084(GSM855970) "background strain: C57BL6/SV129cell type: KR. (dicer cell line) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR206940(GSM723281) other. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM509280(GSM509280) small RNA cloning by length. (testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR065049(SRR065049) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR037912(GSM510449) newborn_rep5. (total RNA) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037920(GSM510458) e12p5_rep2. (embryo) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR206941(GSM723282) other. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR037902(GSM510438) testes_rep3. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................GAGGATGGTGGATGCGTGT.......................................................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ..............................................................................................................................................................................TTCCTGTCCTTTGCCCGGC................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................GAGGATGGTGGATGCGTG........................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................GGCAAAGCAGAGGGCACCGCC.................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......TGAGCGGGCTGACTC........................................................................................................................................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ................................................................................................................................................................................................AAGGCGCCAACATC................................... | 14 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |