| (1) AGO.mut | (5) AGO2.ip | (1) AGO3.ip | (10) BRAIN | (3) CELL-LINE | (1) DGCR8.mut | (9) EMBRYO | (6) ESC | (3) HEART | (2) KIDNEY | (9) LIVER | (1) LUNG | (10) OTHER | (7) OTHER.mut | (2) PIWI.ip | (1) PIWI.mut | (2) SKIN | (2) SPLEEN | (16) TESTES | (1) THYMUS | (5) TOTAL-RNA |
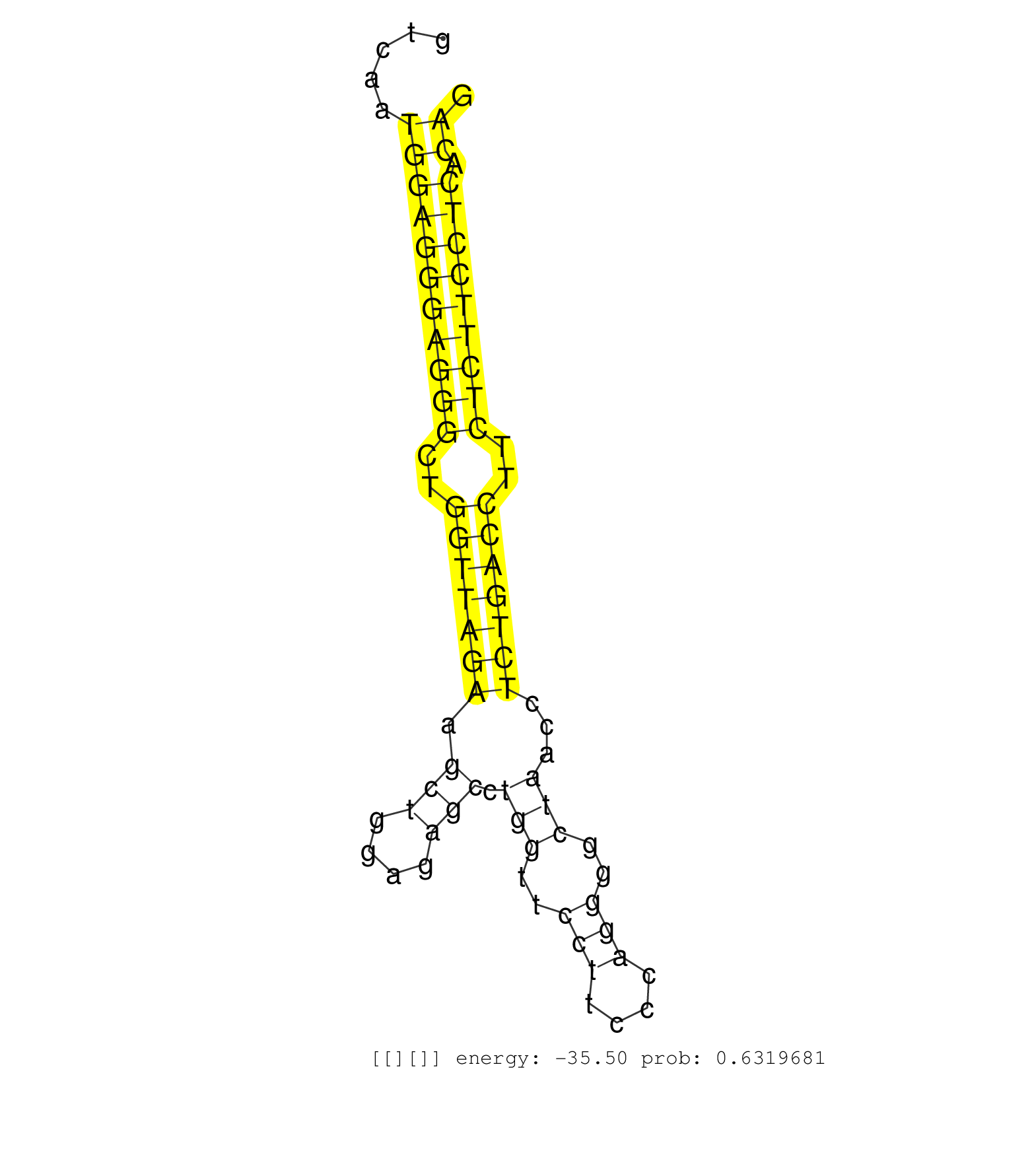
| CCCCTGAGGGCTACTTTGGGAATATCTATAGCACCCCACTGGCACTACAGGTAAGAGAGAGACTCTGGAGCCATGACTGCTTAGCGGGTCAATGGAGGGAGGGCTGGTTAGAAGCTGGAGAGCCTGGTTCCTTCCCAGGGGCTAACCTCTGACCTTCTCTTCCTCACAGATGCTCATGACATCTCCTGCATCCGGGGTAGGGCTGGGCACAGCGTGCAT ............................................................................................(((((((((((..(((((((.(((....))).(((..(((....)))..)))...)))))))..))))))))).))................................................... .......................................................................................88...............................................................................169................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | mjLiverWT3() Liver Data. (liver) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | mjTestesWT4() Testes Data. (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | mjTestesWT1() Testes Data. (testes) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR553585(SRX182791) source: Kidney. (Kidney) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR073954(GSM629280) total RNA. (blood) | SRR206942(GSM723283) other. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR553586(SRX182792) source: Testis. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR095855BC2(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR037912(GSM510449) newborn_rep5. (total RNA) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR095855BC1(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR037914(GSM510451) newborn_rep7. (total RNA) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR553584(SRX182790) source: Heart. (Heart) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR059765(GSM562827) DN3_control. (thymus) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR037910(GSM510447) newborn_rep3. (total RNA) | SRR037902(GSM510438) testes_rep3. (testes) | SRR037930(GSM510468) e7p5_rep4. (embryo) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR065050(SRR065050) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................TCTGACCTTCTCTTCCTCACAGT................................................. | 23 | 1 | 17.00 | 4.00 | 2.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGA........................................................................................................... | 20 | 1 | 15.00 | 15.00 | - | - | 5.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ATGGAGGGAGGGCTGGTTAGA........................................................................................................... | 21 | 1 | 13.00 | 13.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | 4.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....GAGGGCTACTTTGGGAAT.................................................................................................................................................................................................... | 18 | 1 | 9.00 | 9.00 | - | - | - | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAA.......................................................................................................... | 21 | 1 | 9.00 | 9.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCTGACCTTCTCTTCCTCAC.................................................... | 20 | 1 | 7.00 | 7.00 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAG......................................................................................................... | 22 | 1 | 7.00 | 7.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAA......................................................................................................... | 22 | 1 | 5.00 | 9.00 | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGACCTTCTCTTCCTCACAGT................................................. | 21 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................TCTGACCTTCTCTTCCTCACAG.................................................. | 22 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ATGGAGGGAGGGCTGGTTAG............................................................................................................ | 20 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAGT........................................................................................................ | 23 | 1 | 3.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAG............................................................................................................ | 19 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................CTCTGACCTTCTCTTCCTCACA................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................TGGGAATATCTATAGCACCCCACTGGCAC.............................................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..CCTGAGGGCTACTTTGGGAA..................................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................GGTAGGGCTGGGCACAGC...... | 18 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCTGACCTTCTCTTCCTCACAGA................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................ATCCGGGGTAGGGCTGGGCACAGCGTG... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................GCATCCGGGGTAGGGCTGGGCACAGCGTG... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................AATGGAGGGAGGGCTGGTTAGAA.......................................................................................................... | 23 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GGAGGGAGGGCTGGTTAGA........................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGAGAGAGACTCTGGAGCCATGA............................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGT........................................................................................................... | 20 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............TTTGGGAATATCTATAG............................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCCGGGGTAGGGCTGGGCACAGCGT.... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................CATCCGGGGTAGGGCTGGGCACAGCGTG... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CTGAGGGCTACTTTGGGAAT.................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAGTAT...................................................................................................... | 25 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CATCTCCTGCATCCGGGGTAGGGCTGG............. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGGCTGGGCACAGCGCCC.. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................TCTGACCTTCTCTTCCTCACA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAGTT....................................................................................................... | 24 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCTGACCTTCTCTTCCTCACTGA................................................. | 23 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAAT........................................................................................................ | 23 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CCTGCATCCGGGGTAG................... | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AGGGAGGGCTGGTTAGAAGCTT...................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................CATGACATCTCCTGCATCCGGGGTAGGGC................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TGCATCCGGGGTAGGGC................ | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CCCCACTGGCACTACCGCT....................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................TAGAAGCTGGAGAGCCCTA............................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ..........................................................................................AATGGAGGGAGGGCTGGTTAGA........................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GGGCTACTTTGGGAATATC................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................ACAGATGCTCATGACATCTCCTGC.............................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GGAGGGAGGGCTGGTTAGAAGCT....................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TGAGGGCTACTTTGGGAATATCT................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGGGAATATCTATAGCACCCCACTGGCACT............................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................GCATCCGGGGTAGGGCTGG............. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCCGGGGTAGGGCTGGGCAC......... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GGCTACTTTGGGAATATCTATAGCACCCC...................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ATGGAGGGAGGGCTGGTTAGTTT......................................................................................................... | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TCCTGCATCCGGGGTAGGGCTGGGC........... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................ACAGATGCTCATGACATCTCCT................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAGC........................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGTT.......................................................................................................... | 21 | 1 | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TAACCTCTGACCTTCTCTTCCTA...................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................CTCTGACCTTCTCTTCCTCAC.................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................ATCCGGGGTAGGGCTGGGCACAGCGTGC.. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TTGGGAATATCTATAGC........................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ATGGAGGGAGGGCTGGTTAGAAGT........................................................................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAGA........................................................................................................ | 23 | 1 | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................GGGTAGGGCTGGGCACAGCGTGCA. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAT.......................................................................................................... | 21 | 1 | 1.00 | 15.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TTCCCAGGGGCTAACCTCCG.................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................CTCTGACCTTCTCTTCCTCACATA................................................. | 24 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAGGGAGGGCTGGTTAGAAT......................................................................................................... | 22 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CGGGGTAGGGCTGGGCACAGCGTA... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ......................................................................................................................................................................................................AGGGCTGGGCACAGCGTGCAT | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................GGTAGGGCTGGGCACAG....... | 17 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GGCTAACCTCTGACC................................................................. | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CGGGGTAGGGCTGGG............ | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| ................................................................................................................................................................................................CGGGGTAGGGCTGGGTCG......... | 18 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCCTGAGGGCTACTTTGGGAATATCTATAGCACCCCACTGGCACTACAGGTAAGAGAGAGACTCTGGAGCCATGACTGCTTAGCGGGTCAATGGAGGGAGGGCTGGTTAGAAGCTGGAGAGCCTGGTTCCTTCCCAGGGGCTAACCTCTGACCTTCTCTTCCTCACAGATGCTCATGACATCTCCTGCATCCGGGGTAGGGCTGGGCACAGCGTGCAT ............................................................................................(((((((((((..(((((((.(((....))).(((..(((....)))..)))...)))))))..))))))))).))................................................... .......................................................................................88...............................................................................169................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | mjLiverWT3() Liver Data. (liver) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | mjTestesWT4() Testes Data. (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | mjTestesWT1() Testes Data. (testes) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR553585(SRX182791) source: Kidney. (Kidney) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR073954(GSM629280) total RNA. (blood) | SRR206942(GSM723283) other. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR553586(SRX182792) source: Testis. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR095855BC2(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR037912(GSM510449) newborn_rep5. (total RNA) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR095855BC1(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR037914(GSM510451) newborn_rep7. (total RNA) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR553584(SRX182790) source: Heart. (Heart) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR059765(GSM562827) DN3_control. (thymus) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR037910(GSM510447) newborn_rep3. (total RNA) | SRR037902(GSM510438) testes_rep3. (testes) | SRR037930(GSM510468) e7p5_rep4. (embryo) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR065050(SRR065050) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................CCTGGTTCCTTCCCATAG............................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........CTACTTTGGGAATATCT................................................................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................GGAGCCATGACTGCTCGC....................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................TGGAGCCATGACTGCAA......................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |