| (6) AGO2.ip | (4) B-CELL | (21) BRAIN | (2) CELL-LINE | (8) EMBRYO | (3) ESC | (2) FIBROBLAST | (3) LIVER | (1) LUNG | (2) LYMPH | (9) OTHER | (1) PIWI.ip | (2) PIWI.mut | (1) SPLEEN | (9) TESTES | (3) THYMUS | (2) TOTAL-RNA |
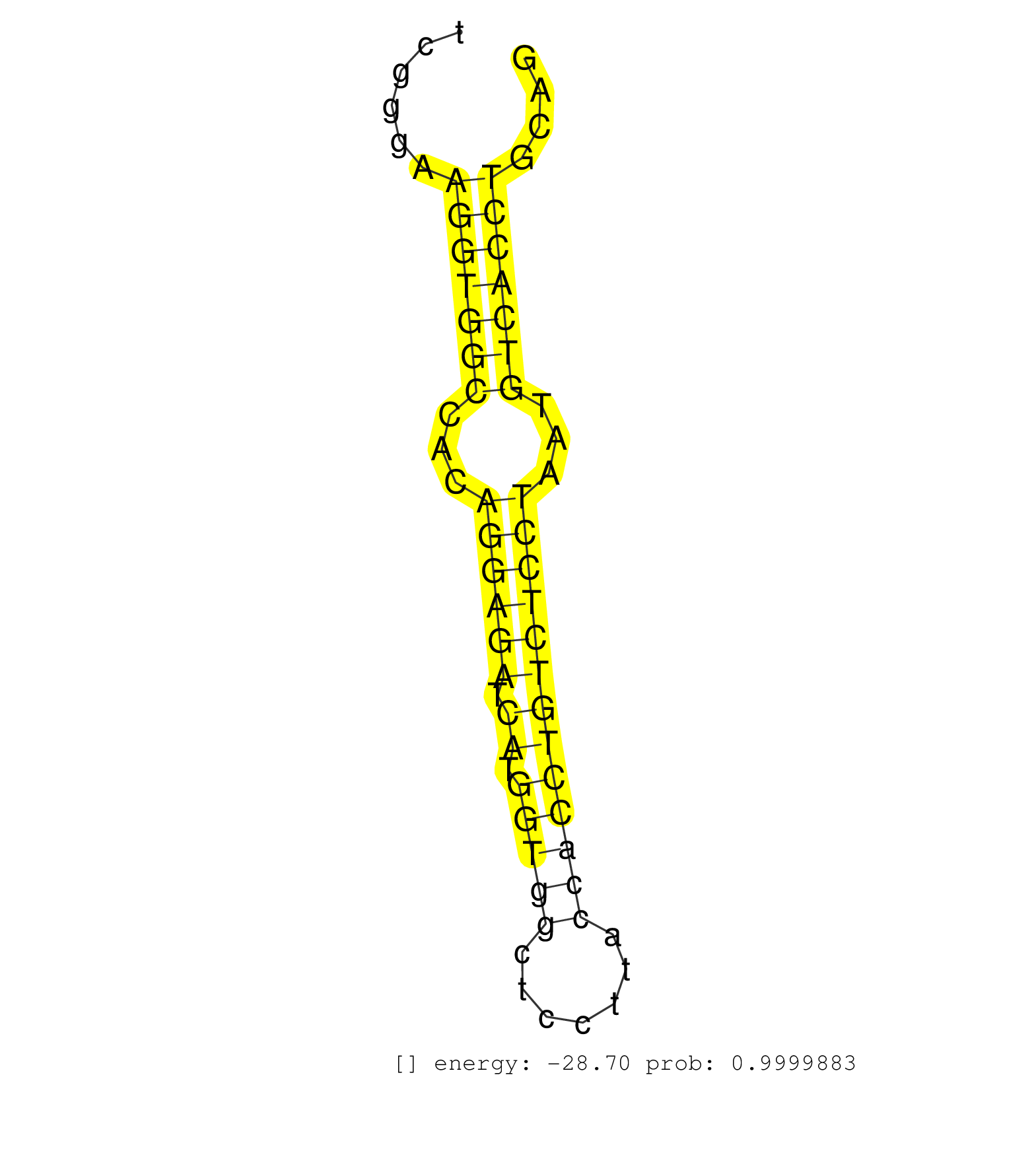
| CGCCAGGGTTGTGTGCAGCCTGACTGTCACTCGTGAATGAGTGACAGTGGCTTCCCACTCTCTCCCTATCCATTAAAGGCCGGGAACTAGTGTGCATTTCCTACACGGGCCACTAGACTAAGGAAGATTATACCCTCGGGAAGGTGGCCACAGGAGATCATGGTGGCTCCTTACCACCTGTCTCCTAATGTCACCTGCAGTTTCTTCCTGCGCCTCTGTTCAGAGGTCCCCATTCTGGAGGACACGCTGA .............................................................................................................................................(((((((...((((((.((.(((((.......)))))))))))))...)))))))...................................................... .......................................................................................................................................136.............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM640577(GSM640577) small RNA in the liver with paternal control . (liver) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR206940(GSM723281) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | GSM640583(GSM640583) small RNA in the liver with paternal Low prot. (liver) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR206939(GSM723280) other. (brain) | SRR037907(GSM510443) brain_rep4. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR073954(GSM629280) total RNA. (blood) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR553582(SRX182788) source: Brain. (Brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR037906(GSM510442) brain_rep3. (brain) | GSM416732(GSM416732) MEF. (cell line) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................AAGGTGGCCACAGGAGATCATGGT...................................................................................... | 24 | 1 | 22.00 | 22.00 | 1.00 | 1.00 | 1.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCATG........................................................................................ | 22 | 1 | 11.00 | 11.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCATGG....................................................................................... | 23 | 1 | 10.00 | 10.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCATGGTGT.................................................................................... | 26 | 1 | 9.00 | 5.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCATGGTG..................................................................................... | 25 | 1 | 5.00 | 5.00 | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................CCTGTCTCCTAATGTCACCTGCAG.................................................. | 24 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CCTGTCTCCTAATGTCACCTGT.................................................... | 22 | 1 | 3.00 | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CCTGTCTCCTAATGTCACCTGC.................................................... | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCAT......................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGGTGGCCACAGGAGATCATGGTGT.................................................................................... | 25 | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TGTCTCCTAATGTCACCTGCAGA................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CCTGTCTCCTAATGTCACCT...................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................TTCTGGAGGACACGCTGA | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCATGGA...................................................................................... | 24 | 1 | 2.00 | 10.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGTCTCCTAATGTCACCTGCAG.................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCA.......................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GTTCAGAGGTCCCCATTCTGGAGGAC....... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CCTGTCTCCTAATGTCACCTACA................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CCTGTCTCCTAATGTCACCTGCA................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................AATGTCACCTGCAGTTTCTTCCT......................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CTCCTAATGTCACCTGCAG.................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CCTGTCTCCTAATGTCACCTGCAT.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................AGGAAGATTATACCCCATA............................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCATGA....................................................................................... | 23 | 1 | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TCTGGAGGACACGCTTTT | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................AATGTCACCTGCAGTTTCTT............................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................CATTCTGGAGGACACGCT.. | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................TTCTGGAGGACACGCAGAG | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................CCTGTCTCCTAATGTCACCTG..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AAGGTGGCCACAGGAGATCATGGTGTAAT................................................................................. | 29 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................ATACCCTCGGGAAGGAGAA...................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................TTCTGGAGGACACGCTG. | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TCTGGAGGACACGCTGCTGA | 20 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................ACGGGCCACTAGACTAAGGAAGATT......................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CTAATGTCACCTGCAAT................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....AGGGTTGTGTGCAGCCTCTCT................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................CTGTCTCCTAATGTCACCTGCA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GTGAATGAGTGACAGTGGCT...................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................ATTCTGGAGGACACG.... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ..........................................................................AAAGGCCGGGAACT.................................................................................................................................................................. | 14 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CGCCAGGGTTGTGTGCAGCCTGACTGTCACTCGTGAATGAGTGACAGTGGCTTCCCACTCTCTCCCTATCCATTAAAGGCCGGGAACTAGTGTGCATTTCCTACACGGGCCACTAGACTAAGGAAGATTATACCCTCGGGAAGGTGGCCACAGGAGATCATGGTGGCTCCTTACCACCTGTCTCCTAATGTCACCTGCAGTTTCTTCCTGCGCCTCTGTTCAGAGGTCCCCATTCTGGAGGACACGCTGA .............................................................................................................................................(((((((...((((((.((.(((((.......)))))))))))))...)))))))...................................................... .......................................................................................................................................136.............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM640577(GSM640577) small RNA in the liver with paternal control . (liver) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR206940(GSM723281) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | GSM640583(GSM640583) small RNA in the liver with paternal Low prot. (liver) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR206939(GSM723280) other. (brain) | SRR037907(GSM510443) brain_rep4. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR073954(GSM629280) total RNA. (blood) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR553582(SRX182788) source: Brain. (Brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR037906(GSM510442) brain_rep3. (brain) | GSM416732(GSM416732) MEF. (cell line) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................CTAATGTCACCTGCAGT................................................. | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................TCTTCCTGCGCCTCTGTG.............................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................TTTCTTCCTGCGCCTAACA............................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................TACACGGGCCACTA....................................................................................................................................... | 14 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |