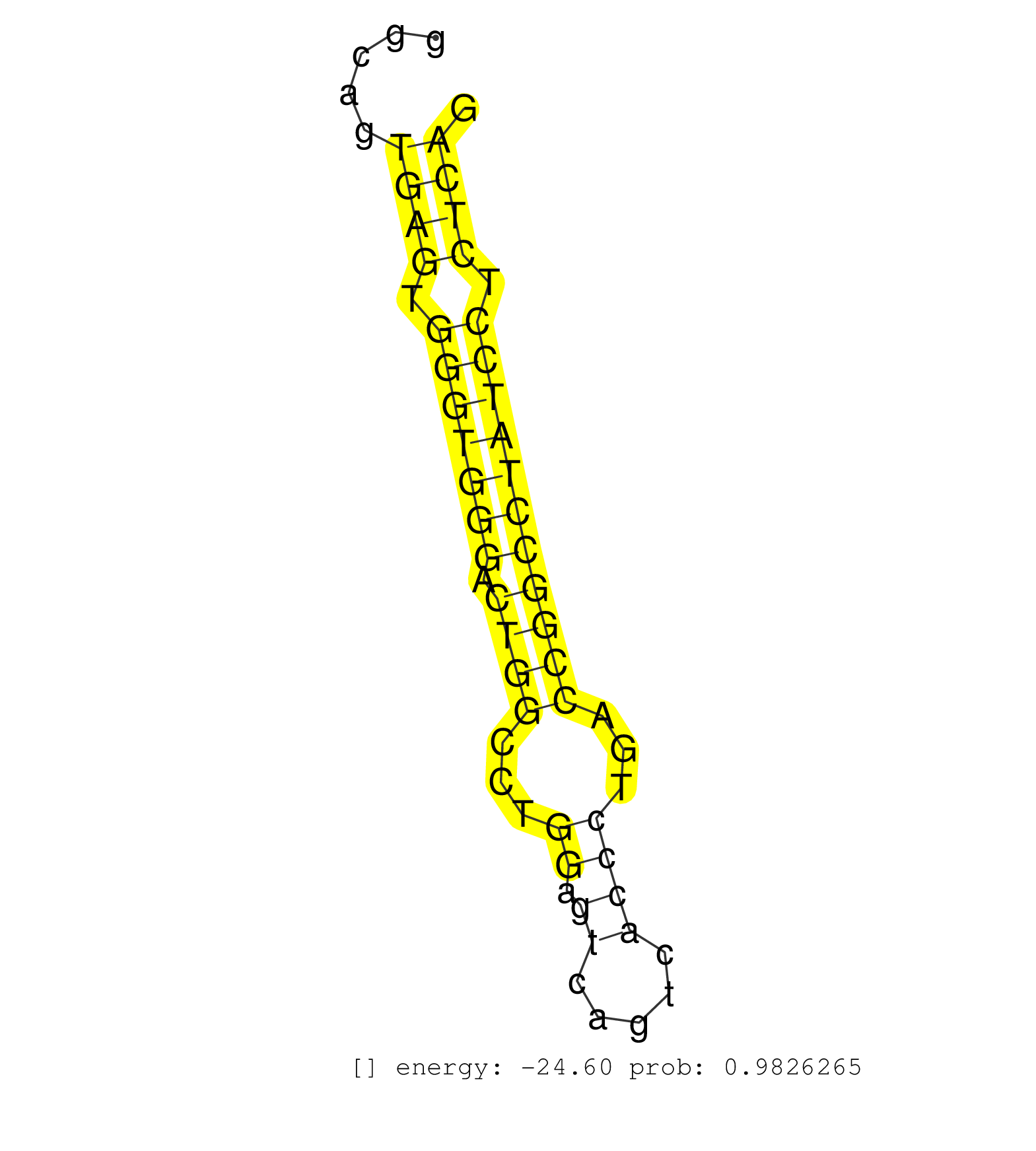
| (1) AGO.mut | (3) AGO1.ip | (11) AGO2.ip | (2) AGO3.ip | (12) B-CELL | (21) BRAIN | (5) CELL-LINE | (3) DCR.mut | (2) DGCR8.mut | (12) EMBRYO | (10) ESC | (5) FIBROBLAST | (6) HEART | (3) KIDNEY | (12) LIVER | (1) LUNG | (5) LYMPH | (26) OTHER | (7) OTHER.mut | (2) OVARY | (2) PANCREAS | (1) PIWI.ip | (2) PIWI.mut | (7) SKIN | (7) SPLEEN | (21) TESTES | (2) THYMUS | (6) TOTAL-RNA | (3) UTERUS |
| CCCTGCGGCGCTGCCAGCAAGAGGGCCAGGAGCTGCTGCGCCACAACCAGGTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGCCTATCCTCTCAGGAGCTGCACTCCCACCTGTCCGAGGAGATCGACCGTCTGCGCAGCTTCAT .........................................................................((((.(((((((.((((...((.((.....))))...))))))))))).))))................................................... ....................................................................69........................................................127................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | mjTestesWT1() Testes Data. (testes) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | mjTestesWT3() Testes Data. (testes) | mjLiverKO1() Liver Data. (Zcchc11 liver) | mjLiverWT1() Liver Data. (liver) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | mjLiverWT2() Liver Data. (liver) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM314559(GSM314559) ESC dgcr8 (454). (ESC) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR059769(GSM562831) Treg_Drosha. (spleen) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR553585(SRX182791) source: Kidney. (Kidney) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR073954(GSM629280) total RNA. (blood) | mjLiverWT3() Liver Data. (liver) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR060845(GSM561991) total RNA. (brain) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR059768(GSM562830) Treg_control. (spleen) | SRR095855BC5(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR394084(GSM855970) "background strain: C57BL6/SV129cell type: KR. (dicer cell line) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR037905(GSM510441) brain_rep2. (brain) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR065052(SRR065052) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR065049(SRR065049) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069834(GSM304914) Analysis of small RNAs in murine neutrophils cultured in vitro by Solexa/Illumina genome analyzer. (blood) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR553584(SRX182790) source: Heart. (Heart) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR525242(SRA056111/SRX170318) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | RuiDGCR8KO(Rui) DGCR8-dependent microRNA biogenesis is essent. (dgcr8 skin) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR095855BC3(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR553603(SRX182809) source: Spermatids. (Spermatids) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR095855BC4(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR553586(SRX182792) source: Testis. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | GSM510444(GSM510444) brain_rep5. (brain) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037929(GSM510467) e7p5_rep3. (embryo) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | GSM475281(GSM475281) total RNA. (testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR346415(SRX098256) source: Testis. (Testes) | SRR206941(GSM723282) other. (brain) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR037911(GSM510448) newborn_rep4. (total RNA) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................TGACCGGCCTATCCTCTCAGT................................................. | 21 | 1 | 461.00 | 41.00 | 86.00 | 24.00 | - | 20.00 | 15.00 | 21.00 | 13.00 | 4.00 | 12.00 | 4.00 | 2.00 | 7.00 | 6.00 | 4.00 | 6.00 | 5.00 | 6.00 | 7.00 | 2.00 | 7.00 | - | 5.00 | 4.00 | 6.00 | 6.00 | 6.00 | 8.00 | 3.00 | 8.00 | 2.00 | 2.00 | 7.00 | 1.00 | 6.00 | - | 1.00 | 1.00 | 4.00 | 2.00 | 2.00 | 4.00 | 5.00 | 5.00 | 3.00 | 2.00 | 3.00 | 1.00 | - | 2.00 | - | 2.00 | 3.00 | 4.00 | 3.00 | - | 2.00 | 4.00 | 1.00 | 2.00 | 3.00 | 2.00 | 2.00 | 3.00 | 3.00 | 1.00 | 1.00 | - | 2.00 | 2.00 | 2.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | 3.00 | - | - | 2.00 | - | 1.00 | 1.00 | 2.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 2.00 | 2.00 | 1.00 | - | 2.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | 2.00 | 1.00 | 2.00 | 1.00 | - | - | - | 2.00 | 2.00 | - | - | 2.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGA................................................. | 21 | 1 | 131.00 | 41.00 | 11.00 | 16.00 | - | 3.00 | 1.00 | 2.00 | 5.00 | 1.00 | 7.00 | 2.00 | 14.00 | 9.00 | - | 8.00 | 1.00 | 1.00 | 3.00 | 1.00 | 4.00 | - | - | 1.00 | - | 1.00 | - | 2.00 | - | 3.00 | - | 1.00 | 2.00 | - | 3.00 | - | - | 1.00 | 1.00 | - | 3.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAG.................................................. | 20 | 1 | 41.00 | 41.00 | 10.00 | - | - | - | - | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTT................................................ | 22 | 1 | 37.00 | 41.00 | 9.00 | - | - | 2.00 | 12.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGCCTATCCTCTCAG.................................................. | 77 | 1 | 23.00 | 23.00 | - | - | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGC................................................. | 21 | 1 | 16.00 | 41.00 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGCCTGGAGTCAGTCACC....................................................................... | 18 | 1 | 13.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTA................................................ | 22 | 1 | 13.00 | 41.00 | - | 1.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | 3.00 | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGAGTGGGTGGGACTGGCCTGG.................................................................................. | 22 | 1 | 12.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTTT............................................... | 23 | 1 | 10.00 | 41.00 | 6.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................AGTGAGTGGGTGGGACTGGCCTGGTT................................................................................ | 26 | 1 | 9.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGAG................................................ | 22 | 1 | 8.00 | 41.00 | - | - | - | - | - | - | - | - | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTAT............................................... | 23 | 1 | 7.00 | 41.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGAA................................................ | 22 | 1 | 7.00 | 41.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCA................................................... | 19 | 1 | 6.00 | 6.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCATT................................................. | 21 | 1 | 6.00 | 6.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGCCTATCCTCTCA................................................... | 76 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGAT................................................ | 22 | 1 | 5.00 | 41.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CAGTGAGTGGGTGGGACTGG....................................................................................... | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAT.................................................. | 20 | 1 | 3.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGAGA............................................... | 23 | 1 | 3.00 | 41.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGCCTATCCTCTCAGA................................................. | 78 | 1 | 3.00 | 23.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGCCTATCCTCTCCG.................................................. | 77 | 3.00 | 0.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................AGTGAGTGGGTGGGACT......................................................................................... | 17 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGCAGGCAGTGAGTGGGTGGGACT......................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGAGTGGGTGGGACTGGCCT.................................................................................... | 20 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGT.............................................................. | 65 | 1 | 3.00 | 2.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GTGAGTGGGTGGGACTGGCCT.................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTTTT.............................................. | 24 | 1 | 2.00 | 41.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGCCTATCCTCTCAGAA................................................ | 79 | 1 | 2.00 | 23.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGTCCGAGGAGATCGACCGTCTGCGC........ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGAAA............................................... | 23 | 1 | 2.00 | 41.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTAG............................................... | 23 | 1 | 2.00 | 41.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTCCCACCTGTCCGAGGA........................ | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGAGTGGGTGGGACTGGCCTGGAAAT.............................................................................. | 26 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................TGAGTGGGTGGGACTGGCCTG................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGGCCTGGAGTCAGTCACC....................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGG............................................................... | 64 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAATT................................................ | 22 | 1 | 2.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TCCGAGGAGATCGACCGT.............. | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTTA............................................... | 23 | 1 | 2.00 | 41.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTAA............................................... | 23 | 1 | 2.00 | 41.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCCGT................................................. | 21 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGTT............................................................. | 66 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................AGTGAGTGGGTGGGACTGGCCTGG.................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGG................................................. | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTCT............................................... | 23 | 1 | 1.00 | 41.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GTGAGTGGGTGGGACTGGCCTGG.................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................ACCGGCCTATCCTCTCAGT................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........CGCTGCCAGCAAGAGGGCC...................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTAT................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCATTC................................................ | 22 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCATTAA............................................... | 23 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCATTTT............................................... | 23 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GTGAGTGGGTGGGACTGGCCTGGAAT............................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................CCTGACCGGCCTATCCTCTCAAAA................................................ | 24 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................CAGTGAGTGGGTGGGACT......................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACCTGTCCGAGGAGATCGA.................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GTGAGTGGGTGGGACTGGCCTGGTTT............................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGTTT............................................................ | 67 | 1 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................AGGAGCTGCACTCCCGCGG................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..CTGCGGCGCTGCCAGCAAGAGGGTCA..................................................................................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................GAGATCGACCGTCTGCGCAGCTTC.. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GACCGGCCTATCCTCTCATT................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................GACCGGCCTATCCTCTCAGT................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGGT............................................................. | 66 | 1 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CTGGCAGGCAGTGAGTGGGT............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GAGGAGATCGACCGTCTGCGCAGCT.... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGAAT............................................... | 23 | 1 | 1.00 | 41.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GCAGGCAGTGAGTGGGTGGGACT......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................AAGAGGGCCAGGAGCTGCTG........................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAAAA................................................ | 22 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTC...................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................CAGGCAGTGAGTGGGTGGGACT......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCT..................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TCCGAGGAGATCGACCGTCTGCGCAGC..... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTC.................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCC................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGCAAGAGGGCCAGGAGCTGCTGCGCC....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGGCCTGGAGTCAGTCACA....................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................TGACCGGCCTATCCTCTCAAGA................................................ | 22 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGCAAGAGGGCCAGGAGA................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................GTGAGTGGGTGGGACTGGCCTGA.................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................CAGGAGCTGCTGCGCGTCA.................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................AGGCAGTGAGTGGGTGGGACT......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGTCCGAGGAGATCGACCGTCTGCGCA....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCTAA................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................AGAGGGCCAGGAGCTGCTGCG......................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAATA................................................ | 22 | 1 | 1.00 | 6.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCATA................................................. | 21 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGAC.................................................................. | 61 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGAGTGGGTGGGACTGGCCTGGAAT............................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................GAGGGCCAGGAGCTGCTGCGCCAC..................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................ACCGGCCTATCCTCTCAGA................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGGTT............................................................ | 67 | 1 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GTGAGTGGGTGGGACTGGCCTGAAT................................................................................ | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGAAG............................................... | 23 | 1 | 1.00 | 41.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAAAT................................................ | 22 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAA.................................................. | 20 | 1 | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCGGGGCCCTGGCAGGCAGTG...................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GTGAGTGGGTGGGACT......................................................................................... | 16 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAGTTG............................................... | 23 | 1 | 1.00 | 41.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCAAA................................................. | 21 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTTAT.................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACCGGCCTATCCTCTCGGA................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTAGGCGGGGCCCTGGCA............................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................AGATCGACCGTC............. | 12 | 7 | 0.57 | 0.57 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | 0.14 | 0.14 |
| .....................AGGGCCAGGAGCTGCTG........................................................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AGTGGGTGGGACTGGC...................................................................................... | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCTGCGGCGCTGCCAGCAAGAGGGCCAGGAGCTGCTGCGCCACAACCAGGTAGGCGGGGCCCTGGCAGGCAGTGAGTGGGTGGGACTGGCCTGGAGTCAGTCACCCTGACCGGCCTATCCTCTCAGGAGCTGCACTCCCACCTGTCCGAGGAGATCGACCGTCTGCGCAGCTTCAT .........................................................................((((.(((((((.((((...((.((.....))))...))))))))))).))))................................................... ....................................................................69........................................................127................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | mjTestesWT1() Testes Data. (testes) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | mjTestesWT3() Testes Data. (testes) | mjLiverKO1() Liver Data. (Zcchc11 liver) | mjLiverWT1() Liver Data. (liver) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | mjLiverWT2() Liver Data. (liver) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM314559(GSM314559) ESC dgcr8 (454). (ESC) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR059769(GSM562831) Treg_Drosha. (spleen) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR553585(SRX182791) source: Kidney. (Kidney) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR073954(GSM629280) total RNA. (blood) | mjLiverWT3() Liver Data. (liver) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR060845(GSM561991) total RNA. (brain) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR059768(GSM562830) Treg_control. (spleen) | SRR095855BC5(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR394084(GSM855970) "background strain: C57BL6/SV129cell type: KR. (dicer cell line) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR037905(GSM510441) brain_rep2. (brain) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR065052(SRR065052) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR065049(SRR065049) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069834(GSM304914) Analysis of small RNAs in murine neutrophils cultured in vitro by Solexa/Illumina genome analyzer. (blood) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR553584(SRX182790) source: Heart. (Heart) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR525242(SRA056111/SRX170318) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | RuiDGCR8KO(Rui) DGCR8-dependent microRNA biogenesis is essent. (dgcr8 skin) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR095855BC3(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR553603(SRX182809) source: Spermatids. (Spermatids) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR095855BC4(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR553586(SRX182792) source: Testis. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | GSM510444(GSM510444) brain_rep5. (brain) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037929(GSM510467) e7p5_rep3. (embryo) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | GSM475281(GSM475281) total RNA. (testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR346415(SRX098256) source: Testis. (Testes) | SRR206941(GSM723282) other. (brain) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR037911(GSM510448) newborn_rep4. (total RNA) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................CCTGTCCGAGGAGATCGACCGTCTGCGCA....... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GGGGCCCTGGCAGGCCAT....................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................GAGATCGACCGTCTGCGG........ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................TCGACCGTCTGCGCAAC..... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................CGGGGCCCTGGCAGG........................................................................................................... | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - |
| ........................................................................................................................................................AGATCGACCGTCT............ | 13 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - |