| (1) AGO2.ip | (1) AGO3.ip | (5) BRAIN | (1) DGCR8.mut | (18) EMBRYO | (1) KIDNEY | (5) LIVER | (1) LYMPH | (3) OTHER | (4) OTHER.mut | (2) SKIN | (3) SPLEEN | (7) TESTES | (10) TOTAL-RNA | (1) UTERUS |
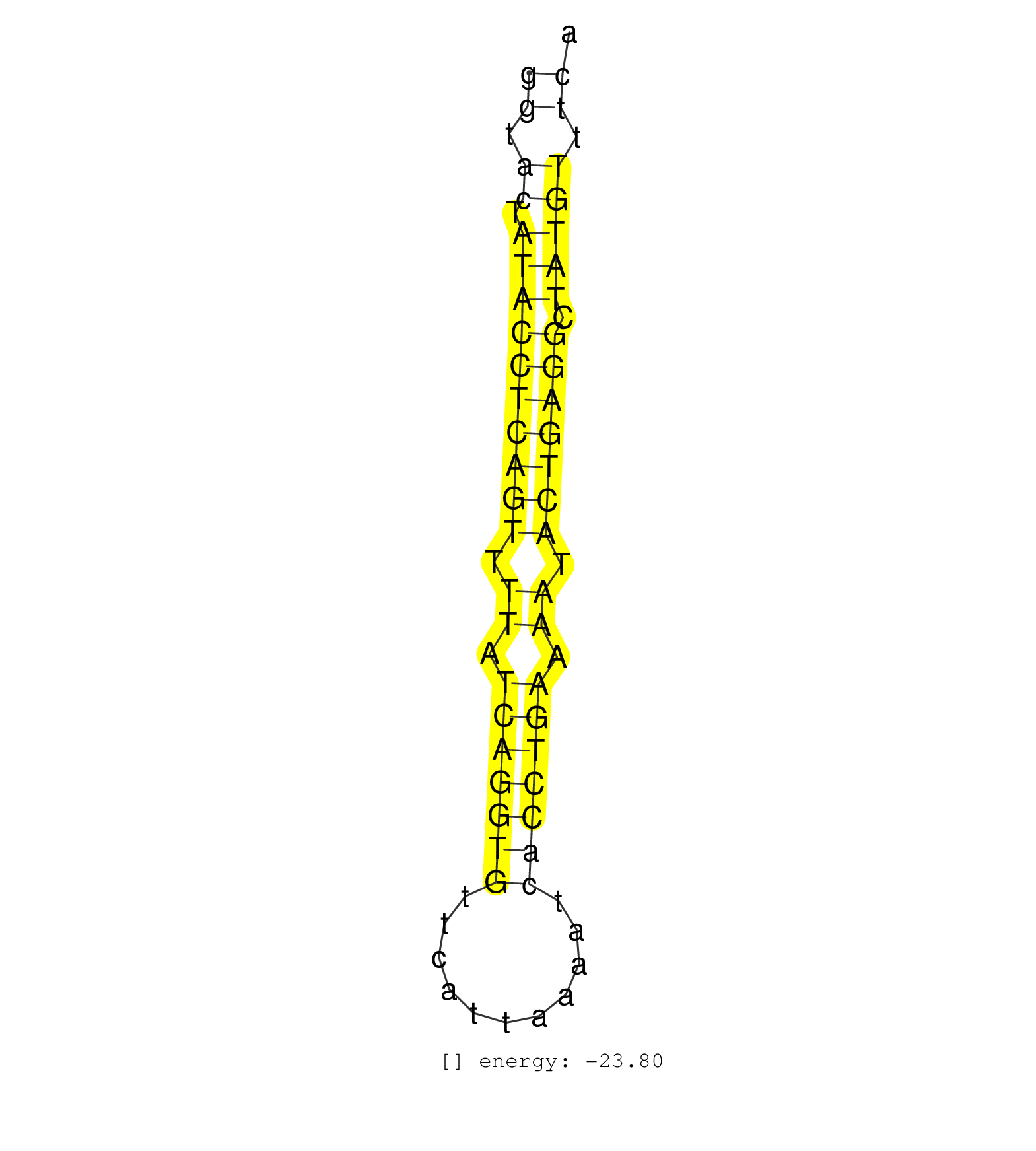
| ATAACCCTGTGCATTTGTTTATTTTTCCTAGGTTTGGGGAAATTCCCAGTTCTGTGGTACTATACCTCAGTTTTATCAGGTGTTCATTAAAATCACCTGAAAATACTGAGGCTATGTTTCACTGAGCATAAGCTTCATGGATTCCTTGAGTGACTGCAGCTGTACTCACCACAAAGAT .......................................................((.((.((((((((((.((.(((((((...........))))))).)).))))))).))))).)).......................................................... .......................................................56...............................................................121....................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037914(GSM510451) newborn_rep7. (total RNA) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM510450(GSM510450) newborn_rep6. (total RNA) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037928(GSM510466) e7p5_rep2. (embryo) | mjLiverWT3() Liver Data. (liver) | SRR037922(GSM510460) e12p5_rep4. (embryo) | SRR037909(GSM510446) newborn_rep2. (total RNA) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR037905(GSM510441) brain_rep2. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR037906(GSM510442) brain_rep3. (brain) | GSM475281(GSM475281) total RNA. (testes) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM317181(GSM317181) total small RNAs from mouse brain. (brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR059769(GSM562831) Treg_Drosha. (spleen) | GSM317182(GSM317182) control IP small RNAs from mouse brain. (brain) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR073954(GSM629280) total RNA. (blood) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037910(GSM510447) newborn_rep3. (total RNA) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR037919(GSM510457) e12p5_rep1. (embryo) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR037903(GSM510439) testes_rep4. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................TATACCTCAGTTTTATCAGGTG................................................................................................ | 22 | 1 | 288.00 | 288.00 | 24.00 | 37.00 | 20.00 | 20.00 | 18.00 | 18.00 | 16.00 | 18.00 | 11.00 | - | 11.00 | 8.00 | 7.00 | 9.00 | 7.00 | 5.00 | 8.00 | 5.00 | 2.00 | 2.00 | 6.00 | 7.00 | - | 1.00 | 5.00 | - | 3.00 | 2.00 | 3.00 | 2.00 | 2.00 | - | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTATGT............................................................. | 22 | 1 | 52.00 | 52.00 | 35.00 | - | - | - | - | - | - | - | - | 7.00 | - | - | 2.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGTGT............................................................................................... | 23 | 1 | 36.00 | 36.00 | 2.00 | 3.00 | 1.00 | 1.00 | 5.00 | 2.00 | 1.00 | - | 1.00 | - | 2.00 | - | - | 2.00 | - | 3.00 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGG.................................................................................................. | 20 | 1 | 27.00 | 27.00 | 1.00 | - | 7.00 | 5.00 | 1.00 | - | - | - | 1.00 | - | - | 2.00 | - | - | 3.00 | - | 1.00 | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGT................................................................................................. | 21 | 1 | 26.00 | 26.00 | - | 1.00 | 3.00 | 1.00 | 4.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | 2.00 | - | 1.00 | 1.00 | - | 3.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGTGa............................................................................................... | 23 | A | 24.00 | 288.00 | 1.00 | 4.00 | 1.00 | - | - | 1.00 | 3.00 | - | 1.00 | - | 2.00 | 4.00 | 1.00 | - | - | 3.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAG................................................................................................... | 19 | 1 | 24.00 | 24.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 7.00 | - | - | - | 1.00 | - | - | - | - | 8.00 | 4.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTATG.............................................................. | 21 | 1 | 7.00 | 7.00 | 5.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CTATACCTCAGTTTTATCAGGTG................................................................................................ | 23 | 1 | 7.00 | 7.00 | - | - | - | - | - | 3.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGTa................................................................................................ | 22 | A | 5.00 | 26.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................TATACCTCAGTTTTATCAGttg................................................................................................ | 22 | TTG | 4.00 | 24.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TCACCTGAAAATACTGAGGCTA................................................................ | 22 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTATGTa............................................................ | 23 | A | 3.00 | 52.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTATGg............................................................. | 22 | G | 2.00 | 7.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAtgtg................................................................................................ | 22 | TGTG | 2.00 | 0.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTAT............................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAtttg................................................................................................ | 22 | TTTG | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTATGaa............................................................ | 23 | AA | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGttgt............................................................................................... | 23 | TTGT | 1.00 | 24.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TCACCTGAAAATACTGAGGCTAT............................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ATACCTCAGTTTTATCAGGTa................................................................................................ | 21 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGcg................................................................................................ | 22 | CG | 1.00 | 27.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTA................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGTGTa.............................................................................................. | 24 | A | 1.00 | 36.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGa................................................................................................. | 21 | A | 1.00 | 27.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTATGTT............................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGTat............................................................................................... | 23 | AT | 1.00 | 26.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTATGTTT........................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ATACCTCAGTTTTATCAGGTG................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................CCTGAAAATACTGAGGCTATt.............................................................. | 21 | T | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGTGaa.............................................................................................. | 24 | AA | 1.00 | 288.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCcggt................................................................................................. | 21 | CGGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TCACCTGAAAATACTGAGGCTt................................................................ | 22 | T | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TATACCTCAGTTTTATCAGGag................................................................................................ | 22 | AG | 1.00 | 27.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ATTCCTTGAGTGACTtggc................... | 19 | TGGC | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ATAACCCTGTGCATTTGTTTATTTTTCCTAGGTTTGGGGAAATTCCCAGTTCTGTGGTACTATACCTCAGTTTTATCAGGTGTTCATTAAAATCACCTGAAAATACTGAGGCTATGTTTCACTGAGCATAAGCTTCATGGATTCCTTGAGTGACTGCAGCTGTACTCACCACAAAGAT .......................................................((.((.((((((((((.((.(((((((...........))))))).)).))))))).))))).)).......................................................... .......................................................56...............................................................121....................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037914(GSM510451) newborn_rep7. (total RNA) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM510450(GSM510450) newborn_rep6. (total RNA) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037928(GSM510466) e7p5_rep2. (embryo) | mjLiverWT3() Liver Data. (liver) | SRR037922(GSM510460) e12p5_rep4. (embryo) | SRR037909(GSM510446) newborn_rep2. (total RNA) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR037905(GSM510441) brain_rep2. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR037906(GSM510442) brain_rep3. (brain) | GSM475281(GSM475281) total RNA. (testes) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM317181(GSM317181) total small RNAs from mouse brain. (brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR059769(GSM562831) Treg_Drosha. (spleen) | GSM317182(GSM317182) control IP small RNAs from mouse brain. (brain) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR073954(GSM629280) total RNA. (blood) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037910(GSM510447) newborn_rep3. (total RNA) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR037919(GSM510457) e12p5_rep1. (embryo) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR037903(GSM510439) testes_rep4. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................GGTTTGGGGAAATTCCC................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |