| (1) AGO2.ip | (9) BRAIN | (6) CELL-LINE | (2) DGCR8.mut | (6) EMBRYO | (1) FIBROBLAST | (1) OTHER | (1) OTHER.mut | (1) PIWI.mut | (4) TESTES |
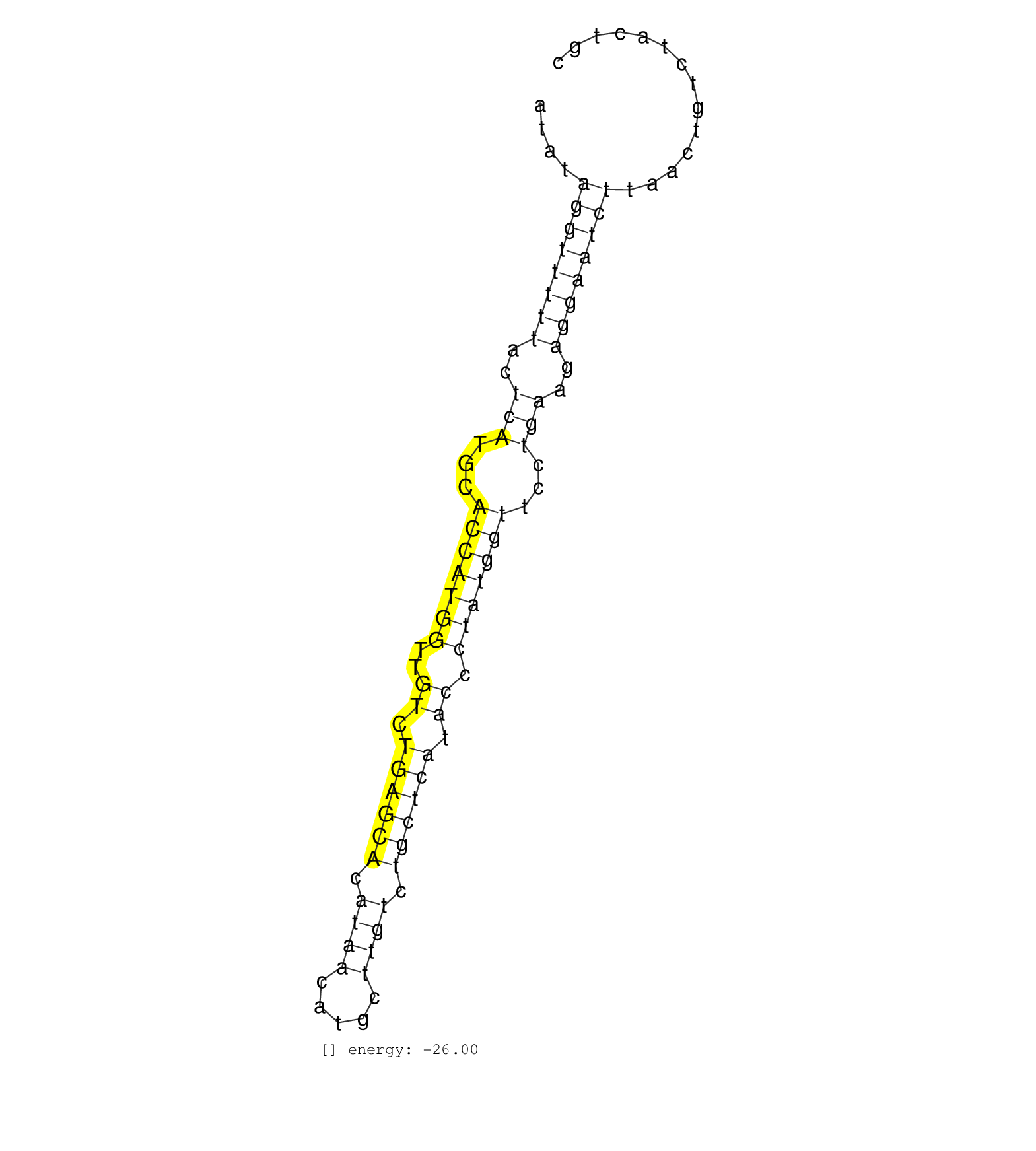
| GGCATATTCCCGTGTTATTTGTTCCTGTATCACATCTACTTTTGTATTATAGAATCCATATAGGTTTTTACTCATGCACCATGGTTGTCTGAGCACATAACATGCTTGTCTGCTCATACCCTATGGTTCCTGAAGAGGAATCTTAACTGTCTACTGCTCTATAGGAAAACAGTGTTTTATGTATCAGTTTATATTTTTGGTTGCTTT ......................................................((((((((..(((...(((((((..((.((((((.((((.....)))).)))))).)).)))))))...)))..))))))))................................................................ .........................................................58.................................................................................................157................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR060845(GSM561991) total RNA. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037937(GSM510475) 293cand2. (cell line) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | GSM566420(GSM566420) Endougenous small RNA from mouse NIH3T3 cell, infected with MHV-68. (fibroblast) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR206940(GSM723281) other. (brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................ATGCACCATGGTTGTCTGAGCA................................................................................................................ | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCAt............................................................................................................... | 22 | T | 4.00 | 1.00 | - | - | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCAtg.............................................................................................................. | 23 | TG | 4.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCAtgc............................................................................................................. | 24 | TGC | 4.00 | 1.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCAtga............................................................................................................. | 24 | TGA | 3.00 | 1.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................ATGCACCATGGTTGTCTGAGC................................................................................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCAta.............................................................................................................. | 23 | TA | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCAa............................................................................................................... | 22 | A | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCA................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCgtga............................................................................................................. | 24 | GTGA | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TTTTATGTATCAGTTggc............... | 18 | GGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCAtac............................................................................................................. | 24 | TAC | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TATAGAATCCATATAac............................................................................................................................................... | 17 | AC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................ATGCTTGTCTGCTCAcgg........................................................................................ | 18 | CGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................ATGCACCATGGTTGTCTGAG.................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCAttc............................................................................................................. | 24 | TTC | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCACCATGGTTGTCTGAGCgt............................................................................................................... | 22 | GT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................AGTTTATATTTTTGGTT..... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| GGCATATTCCCGTGTTATTTGTTCCTGTATCACATCTACTTTTGTATTATAGAATCCATATAGGTTTTTACTCATGCACCATGGTTGTCTGAGCACATAACATGCTTGTCTGCTCATACCCTATGGTTCCTGAAGAGGAATCTTAACTGTCTACTGCTCTATAGGAAAACAGTGTTTTATGTATCAGTTTATATTTTTGGTTGCTTT ......................................................((((((((..(((...(((((((..((.((((((.((((.....)))).)))))).)).)))))))...)))..))))))))................................................................ .........................................................58.................................................................................................157................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR060845(GSM561991) total RNA. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037937(GSM510475) 293cand2. (cell line) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | GSM566420(GSM566420) Endougenous small RNA from mouse NIH3T3 cell, infected with MHV-68. (fibroblast) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR206940(GSM723281) other. (brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................TGCACCATGGTTGTCTGAGCA................................................................................................................ | 21 | 1 | 9.00 | 9.00 | 4.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................aaccTTATATTTTTGGTTG.... | 19 | aacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................aATGCACCATGGTTGTC...................................................................................................................... | 17 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................gaGAGCACATAACATGCTT.................................................................................................... | 19 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................cctgATGGTTGTCTGAGCA................................................................................................................ | 19 | cctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |