| (2) AGO2.ip | (3) BRAIN | (4) CELL-LINE | (9) EMBRYO | (7) FIBROBLAST | (1) LIVER | (2) OTHER | (1) SPLEEN |
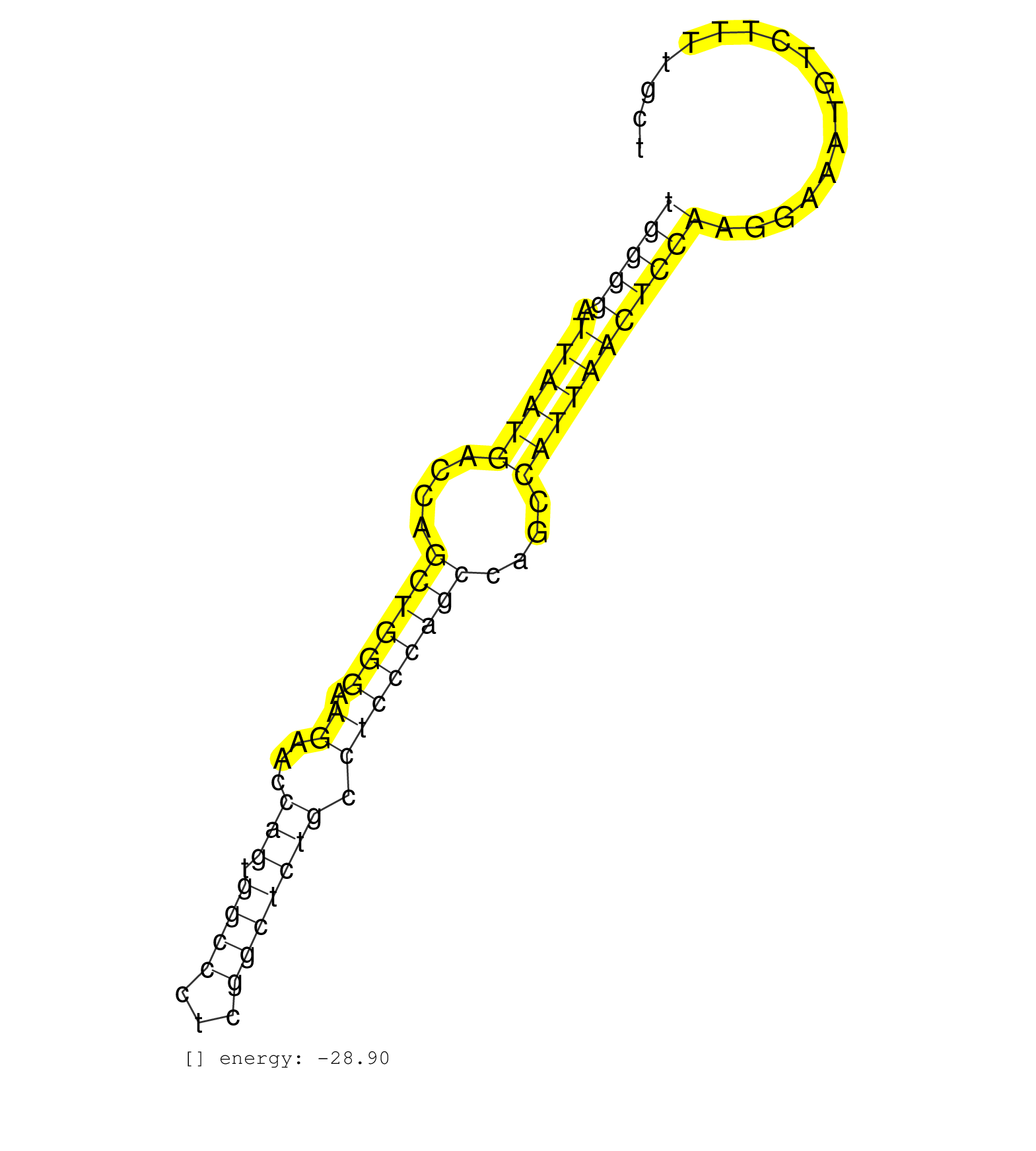
| TTTCCGTACCCTGGAACTGTGAGAAATTTCTGTAATGGTGCTTTAGCAATAATGATGGAGGTGCAGGCGTTTCCTGGGGATTAATGACCAGCTGGGAAGAACCAGTGGCCCTCGGCTCTGCCTCCCAGCCAGCCATTAACTCCAAGGAAATGTCTTTTGCTGAGGTCGTTACTGCTATTATAAGCTTAAAATGGAAAAACATAACAGACATATGAGTTAT ...........................................................................((((.((((((....((((((.((...(((.((((...))))))).))))))))....)))))))))).((.(((.....))).))........................................................... ..........................................................................75....................................................................................161......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR037936(GSM510474) 293cand1. (cell line) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR029123(GSM416611) NIH3T3. (cell line) | GSM416732(GSM416732) MEF. (cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR073954(GSM629280) total RNA. (blood) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM640577(GSM640577) small RNA in the liver with paternal control . (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................ATTAATGACCAGCTGGGAAGAA....................................................................................................................... | 22 | 1 | 24.00 | 24.00 | - | 7.00 | - | 3.00 | 7.00 | - | 1.00 | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAGA........................................................................................................................ | 21 | 1 | 18.00 | 18.00 | 1.00 | 4.00 | - | - | - | - | 1.00 | 3.00 | 2.00 | 3.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAG......................................................................................................................... | 20 | 1 | 13.00 | 13.00 | - | 1.00 | - | - | - | 3.00 | 2.00 | 2.00 | - | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAGAAC...................................................................................................................... | 23 | 1 | 7.00 | 7.00 | - | 2.00 | - | 3.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAGAAa...................................................................................................................... | 23 | A | 7.00 | 24.00 | - | 1.00 | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CAGCCAGCCATTAACTCCAAGt......................................................................... | 22 | T | 6.00 | 0.00 | - | 1.00 | - | 1.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TAGCAATAATGATGGAGGTGC............................................................................................................................................................ | 21 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GCCATTAACTCCAAGGAAATGTCTTT............................................................... | 26 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ACCCTGGAACTGTGAGAAATTTCTGT........................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAGAACC..................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ACCCTGGAACTGTGAGAAATT................................................................................................................................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGGAAGAACCAGTGat............................................................................................................... | 17 | AT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................TGGGAAGAACCAGTGacc.............................................................................................................. | 18 | ACC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................GGAAAAACATAACAGACAT......... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CCTCCCAGCCAGCCATTAACTC.............................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................AAGCTTAAAATGGAAAAACAT.................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................ATTAATGACCAGCTGGGAA.......................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAGAAt...................................................................................................................... | 23 | T | 1.00 | 24.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAGAta...................................................................................................................... | 23 | TA | 1.00 | 18.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TCCCAGCCAGCCATTAACTCCAtt.......................................................................... | 24 | TT | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TGGTGCTTTAGCAATAATGATGGAG................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAGAAg...................................................................................................................... | 23 | G | 1.00 | 24.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................GCCAGCCATTAACTCCAAGGAAATGTCTTT............................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................AAAATGGAAAAACATAACAGACAT......... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTAATGACCAGCTGGGAAaaa....................................................................................................................... | 22 | AAA | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGTAATGGTGCTTTAta............................................................................................................................................................................. | 17 | TA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| TTTCCGTACCCTGGAACTGTGAGAAATTTCTGTAATGGTGCTTTAGCAATAATGATGGAGGTGCAGGCGTTTCCTGGGGATTAATGACCAGCTGGGAAGAACCAGTGGCCCTCGGCTCTGCCTCCCAGCCAGCCATTAACTCCAAGGAAATGTCTTTTGCTGAGGTCGTTACTGCTATTATAAGCTTAAAATGGAAAAACATAACAGACATATGAGTTAT ...........................................................................((((.((((((....((((((.((...(((.((((...))))))).))))))))....)))))))))).((.(((.....))).))........................................................... ..........................................................................75....................................................................................161......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR037936(GSM510474) 293cand1. (cell line) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR029123(GSM416611) NIH3T3. (cell line) | GSM416732(GSM416732) MEF. (cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR073954(GSM629280) total RNA. (blood) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM640577(GSM640577) small RNA in the liver with paternal control . (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................CAAGGAAATGTCTTTTGCT........................................................... | 19 | 1 | 12.00 | 12.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GGAAATGTCTTTTGCTGAGGTCGT................................................... | 24 | 1 | 12.00 | 12.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GGGGATTAATGACCAGCTGGGA........................................................................................................................... | 22 | 1 | 11.00 | 11.00 | 9.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................CTCCCAGCCAGCCATTAACTC.............................................................................. | 21 | 1 | 9.00 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CCTCCCAGCCAGCCATTAACTC.............................................................................. | 22 | 1 | 6.00 | 6.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AAGGAAATGTCTTTTGCTGA......................................................... | 20 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CAAGGAAATGTCTTTTGCTGAGGT...................................................... | 24 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GGGATTAATGACCAGCTGGGA........................................................................................................................... | 21 | 1 | 4.00 | 4.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................aGGGGATTAATGACCAGCTGGGA........................................................................................................................... | 23 | a | 3.00 | 11.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CAAGGAAATGTCTTTTGCTG.......................................................... | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................tCTCCCAGCCAGCCATTAACTC.............................................................................. | 22 | t | 2.00 | 9.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................tCTCCCAGCCAGCCATTAACT............................................................................... | 21 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CCTCCCAGCCAGCCATTAACT............................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CAAGGAAATGTCTTTTGCTGA......................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................tcCCTCCCAGCCAGCCATTAACTC.............................................................................. | 24 | tc | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................CCTCCCAGCCAGCCATTA.................................................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................ctatGAAATGTCTTTTGCT........................................................... | 19 | ctat | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................ccgcACATAACAGACATAT....... | 19 | ccgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................AAGGAAATGTCTTTTGCTG.......................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................aCCTCCCAGCCAGCCATTAACT............................................................................... | 22 | a | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................aCCTCCCAGCCAGCCATTAACTC.............................................................................. | 23 | a | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................tctcGGAGGTGCAGGCGTTT.................................................................................................................................................... | 20 | tctc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................caGCCCTCGGCTCTGCCT................................................................................................. | 18 | ca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................GATTAATGACCAGCTGGGA........................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TCCAAGGAAATGTCTTTTGCTGAGGTCGTTA................................................. | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CAAGGAAATGTCTTTTGC............................................................ | 18 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TCCCAGCCAGCCATTA.................................................................................. | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - |