| (5) BRAIN | (2) CELL-LINE | (1) DGCR8.mut | (3) EMBRYO | (1) ESC | (1) KIDNEY | (1) LYMPH | (4) OTHER | (1) OVARY | (3) TESTES | (1) TOTAL-RNA | (1) UTERUS |
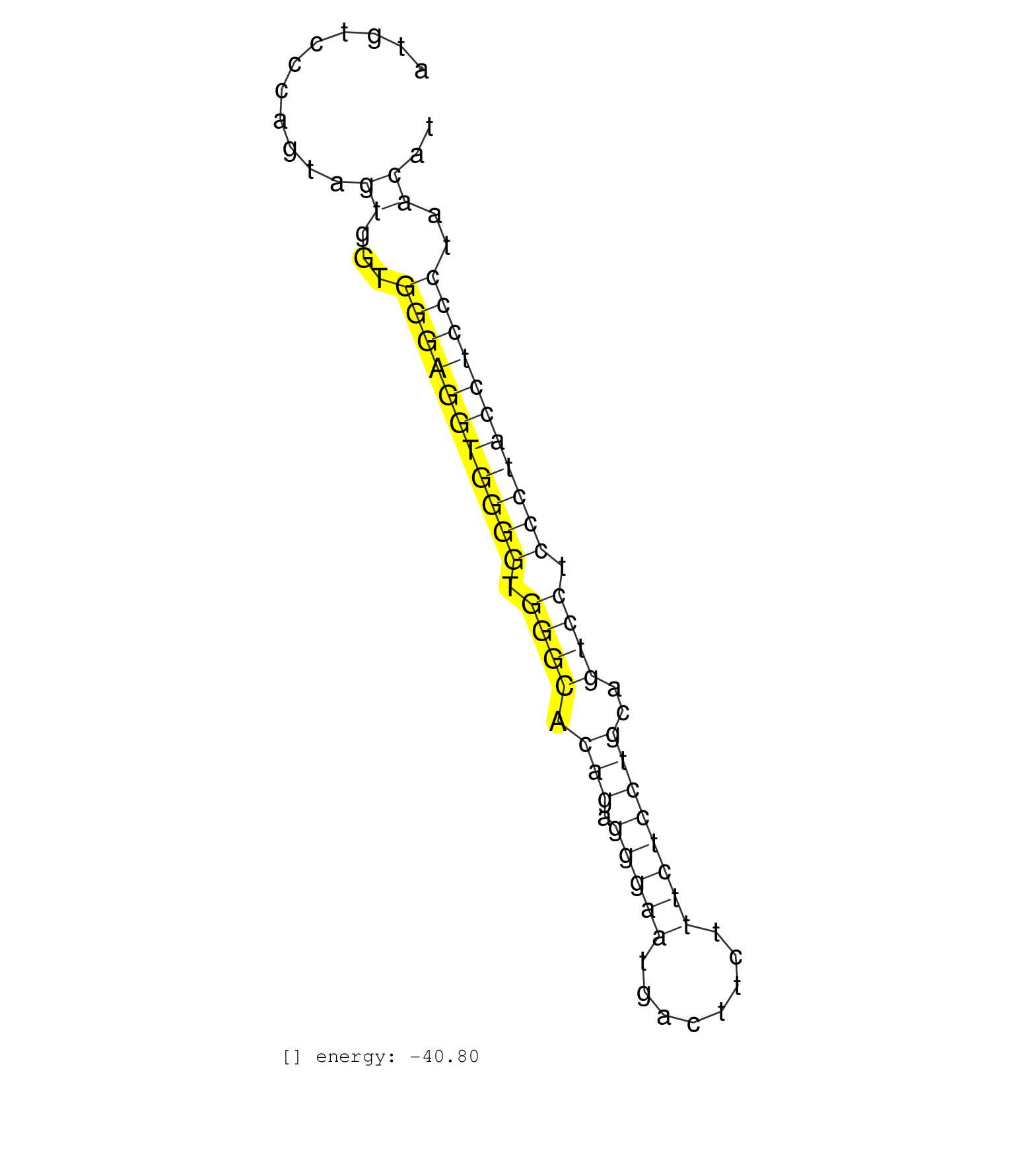
| TGTGCCTATTCCTCAGCTGCCCCCTGCCCCTCTTCAGACCCTCCCACTCCATGTCCCAGTAGTGGTGGGAGGTGGGGTGGGCACAGAGGGAATGACTTCTTTCTCCTGCAGTCCTCCCTACCTCCCTAACATGAAACCCTCCAGGTTGGGGTTTGGGGCTGCTCATGTTTTGTGGACCAGGC .............................................................((...(((((((((((.((((.(((.(((((........))))))))..)))).)))))))))))..)).................................................... ..................................................51...............................................................................132................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR073955(GSM629281) total RNA. (blood) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR073954(GSM629280) total RNA. (blood) | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................ACCCTCCCACTCCATGTCC.............................................................................................................................. | 19 | 2 | 2.50 | 2.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGTGGGGTGGGCACAtttt............................................................................................. | 20 | TTTT | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................AACATGAAACCCTCCAGGTT................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGGTGGGAGGTGGGGTct...................................................................................................... | 18 | CT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGGTTGGGGTTTGGGGCTGCTC.................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TATTCCTCAGCTGCCCCCTGCCCCTCTT.................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............TCAGCTGCCCCCTGCCCCTCTTt................................................................................................................................................... | 23 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GGGGTTTGGGGCTGCccc................. | 18 | CCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................ACCCTCCCACTCCATGccc.............................................................................................................................. | 19 | CCC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCCTGCAGTCCTCCCTACCTCCaag...................................................... | 25 | AAG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GTGGGCACAGAGGGAActtc...................................................................................... | 20 | CTTC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................ATGTTTTGTGGACCAaaa | 18 | AAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................GAAACCCTCCAGGTTGGGGTTTGGGG........................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCCTGCAGTCCTCCCgagc............................................................ | 19 | GAGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................AACATGAAACCCTCCAGGT.................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TCCTGCAGTCCTCCCTACCTCCtaat..................................................... | 26 | TAAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GGTGGGAGGTGGGGTGta..................................................................................................... | 18 | TA | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GTGGGAGGTGGGGTGGGCA................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .....................................................................AGGTGGGGTGGGCACAGA............................................................................................... | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| TGTGCCTATTCCTCAGCTGCCCCCTGCCCCTCTTCAGACCCTCCCACTCCATGTCCCAGTAGTGGTGGGAGGTGGGGTGGGCACAGAGGGAATGACTTCTTTCTCCTGCAGTCCTCCCTACCTCCCTAACATGAAACCCTCCAGGTTGGGGTTTGGGGCTGCTCATGTTTTGTGGACCAGGC .............................................................((...(((((((((((.((((.(((.(((((........))))))))..)))).)))))))))))..)).................................................... ..................................................51...............................................................................132................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR073955(GSM629281) total RNA. (blood) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR073954(GSM629280) total RNA. (blood) | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................CTTCTTTCTCCTGCAGTCCTCCC................................................................ | 23 | 1 | 12.00 | 12.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CTGCCCCTCTTCAGACCCTCCCACTCCATG................................................................................................................................. | 30 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................ttTGAAACCCTCCAGGT.................................... | 17 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ccaaAGGTTGGGGTTTGGGG........................ | 20 | ccaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................tgcaCAGGTTGGGGTTTGGGG........................ | 21 | tgca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................acatACTTCTTTCTCCTGCA........................................................................ | 20 | acat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................cttaTTTCTCCTGCAGTCCTCCC................................................................ | 23 | ctta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................cggcTGGTGGGAGGTGGGG......................................................................................................... | 19 | cggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................ccCCCCTCTTCAGACCCT............................................................................................................................................ | 18 | cc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |