| (1) AGO2.ip | (2) BRAIN | (1) CELL-LINE | (1) EMBRYO | (1) OTHER | (3) OTHER.mut | (1) PIWI.ip | (5) TESTES |
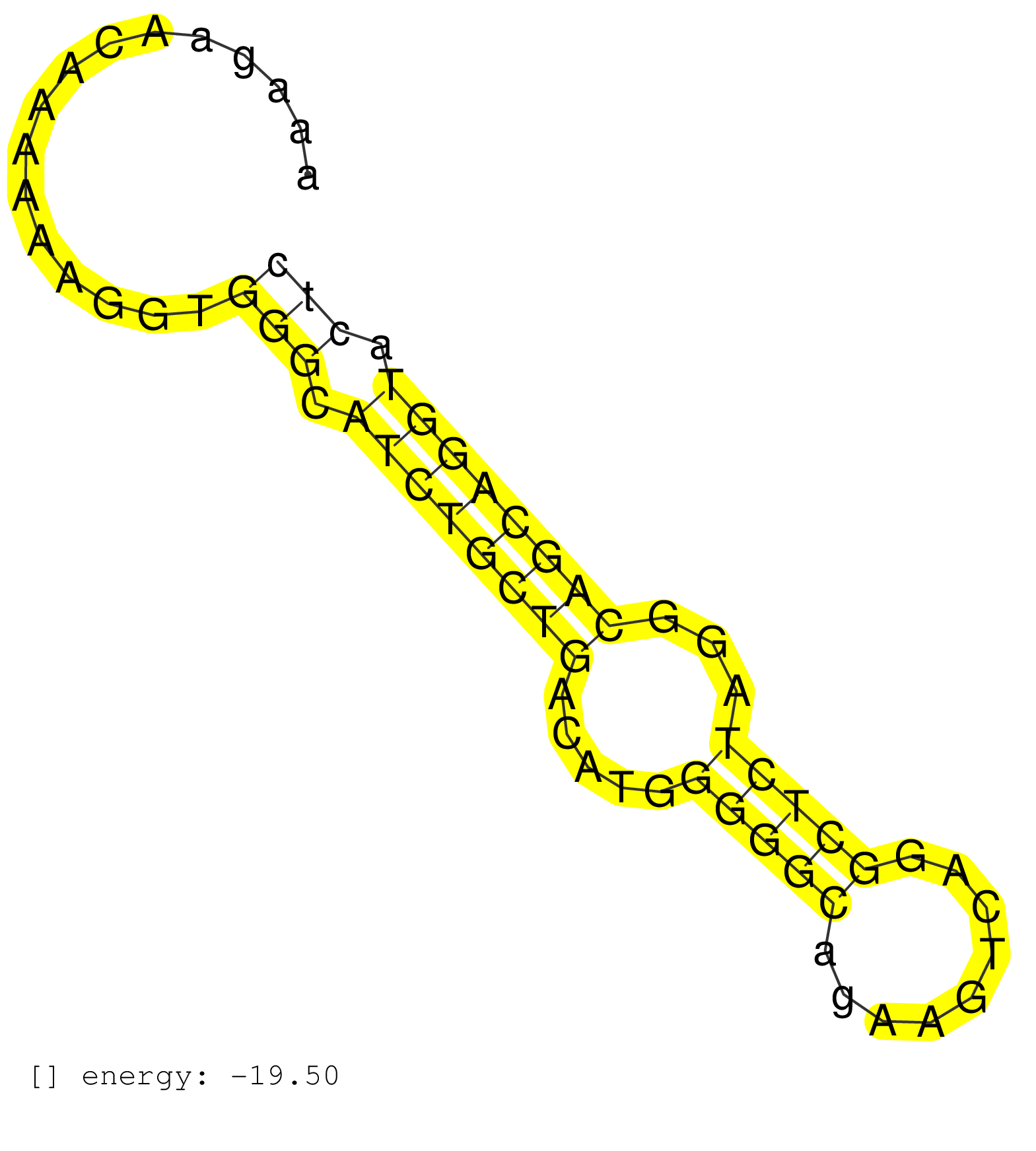
| GAGGTGCCTGGTTTTAAAGCATACATCTCGGGGAATAGGGAGGGGCCGAGGGGGGATTTAACAAAGAACAAAAAAGGTGGGCATCTGCTGACATGGGGGCAGAAGTCAGGCTCTAGGCAGCAGGTACTCTTCATCTTATCTCCAGAACATAGATCCTCCTTAACAGCCTTGGGATATCAGGCCGGGC ..............................................................................(((.((((((((.....(((((.........)))))...)))))))).))).......................................................... ..............................................................63................................................................129........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037936(GSM510474) 293cand1. (cell line) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR073954(GSM629280) total RNA. (blood) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR206942(GSM723283) other. (brain) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............TTAAAGCATACATCTCGGGt.......................................................................................................................................................... | 20 | T | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................AAGTCAGGCTCTAGGCAGCAGGT.............................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................ACAAAAAAGGTGGGCATCTGCTGACATGGGGGC....................................................................................... | 33 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGAACAAAAAAGGcgga.......................................................................................................... | 19 | CGGA | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............TAAAGCATACATCTCGGGGAATAGGa................................................................................................................................................... | 26 | A | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................AATAGGGAGGGGCCGga......................................................................................................................................... | 17 | GA | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................AAGAACAAAAAAGGTGGGC......................................................................................................... | 19 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - |
| GAGGTGCCTGGTTTTAAAGCATACATCTCGGGGAATAGGGAGGGGCCGAGGGGGGATTTAACAAAGAACAAAAAAGGTGGGCATCTGCTGACATGGGGGCAGAAGTCAGGCTCTAGGCAGCAGGTACTCTTCATCTTATCTCCAGAACATAGATCCTCCTTAACAGCCTTGGGATATCAGGCCGGGC ..............................................................................(((.((((((((.....(((((.........)))))...)))))))).))).......................................................... ..............................................................63................................................................129........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037936(GSM510474) 293cand1. (cell line) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR073954(GSM629280) total RNA. (blood) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR206942(GSM723283) other. (brain) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................gcaGTACTCTTCATCTTATCTCCAGAA........................................ | 27 | gca | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................CAGCAGGTACTCTTCATCTTATCTCCA........................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................tcgGGGGCCGAGGGGGGA................................................................................................................................... | 18 | tcg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................gaATCTGCTGACATGGGGG........................................................................................ | 19 | ga | 0.67 | 0.00 | - | - | - | - | - | - | - | - | 0.67 | - |
| .......................................................................................................................................................GATCCTCCTTAACAGCCTTGGGATATCA........ | 28 | 5 | 0.60 | 0.60 | - | - | - | - | - | - | - | - | - | 0.60 |