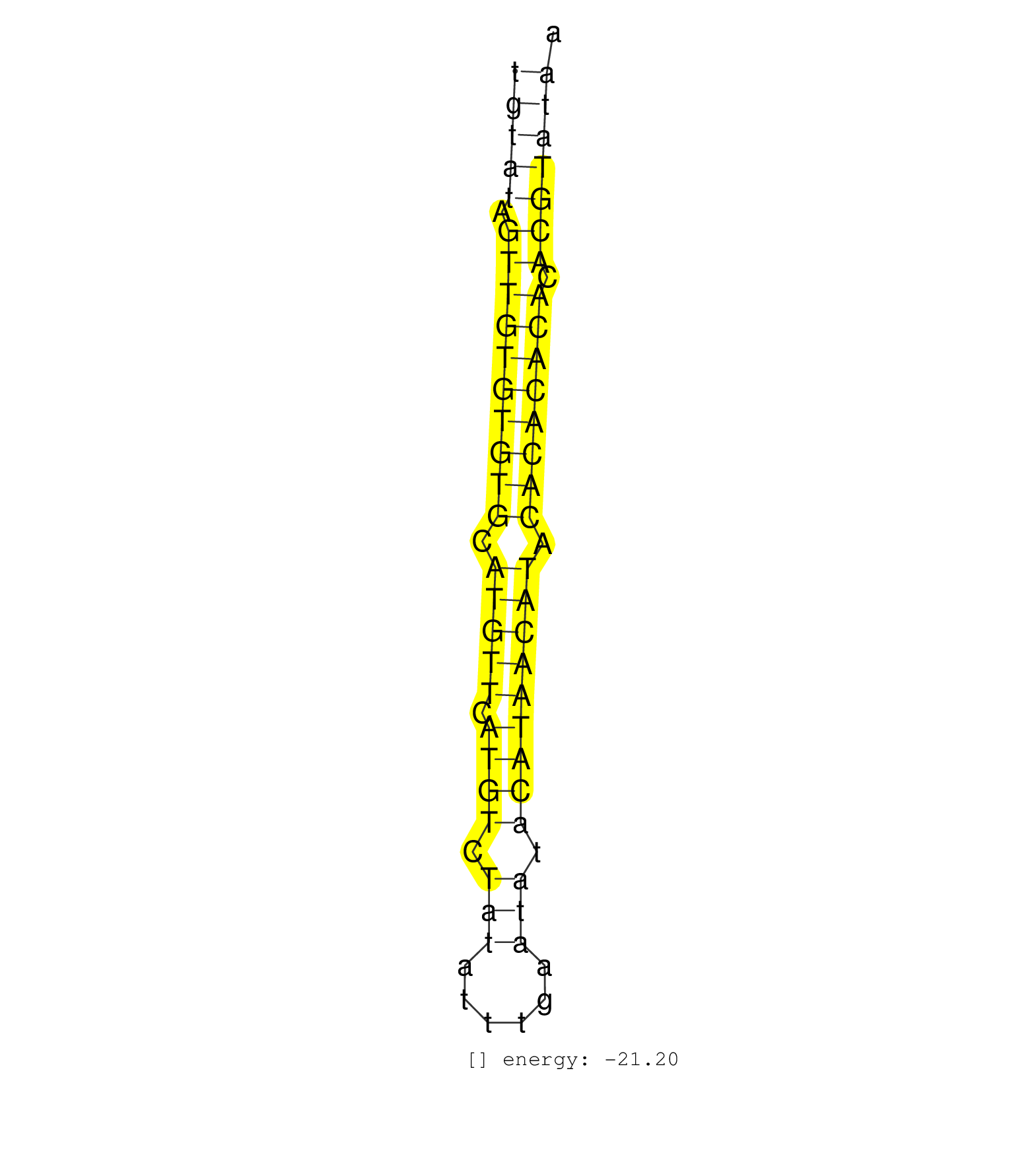
| (1) AGO1.ip | (7) AGO2.ip | (9) B-CELL | (13) BRAIN | (7) CELL-LINE | (1) DGCR8.mut | (12) EMBRYO | (12) ESC | (9) FIBROBLAST | (2) LIVER | (4) LYMPH | (9) OTHER | (10) OTHER.mut | (3) PIWI.ip | (1) PIWI.mut | (1) SKIN | (8) SPLEEN | (26) TESTES | (4) THYMUS | (1) TOTAL-RNA | (2) UTERUS |
| AAATTCATACTTATCCTCATGAGAGATCAGCCTGTGATTCCTCCATGTATGTGCATGTGTGTATAGTTGTGTGTGCATGTTCATGTCTATATTTGAATATACATAACATACACACACACACGTATAAACGCAAGCACACATACACAGACACAGGAATGGCATTCGCCAATCCCTTTGAATGATAAGT ..................................................(((((.((((((((((.(((((.((((.(((......))).))))))))).)))))))).)))))))................................................... ...........................................................60.................................................................127.......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR073954(GSM629280) total RNA. (blood) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR059768(GSM562830) Treg_control. (spleen) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR390298(GSM849856) cell line: NIH-3T3cell type: fibroblastinfect. (fibroblast) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR073955(GSM629281) total RNA. (blood) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | GSM510444(GSM510444) brain_rep5. (brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR037905(GSM510441) brain_rep2. (brain) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM314558(GSM314558) ESC wild type (454). (ESC) | GSM416732(GSM416732) MEF. (cell line) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | GSM261957(GSM261957) oocytesmallRNA-19to24. (oocyte) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR206942(GSM723283) other. (brain) | GSM475281(GSM475281) total RNA. (testes) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | GSM509280(GSM509280) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................CATAACATACACACACACACGT................................................................ | 22 | 2 | 169.00 | 169.00 | 23.00 | 23.00 | - | 5.00 | 12.00 | 18.50 | 1.50 | 1.00 | 10.00 | 1.00 | 4.00 | 3.50 | 1.50 | 1.50 | 4.00 | 0.50 | 5.00 | 2.50 | 2.00 | 3.50 | 3.00 | 3.50 | - | 2.50 | 3.50 | 1.50 | 1.50 | 4.00 | - | 2.00 | 0.50 | - | - | - | 1.50 | 0.50 | 1.00 | 0.50 | 1.50 | 1.00 | - | - | 1.50 | - | - | 0.50 | 0.50 | 0.50 | 1.00 | - | 1.50 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 0.50 | - | - | 0.50 | - | 1.50 | 0.50 | - | - | - | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | 0.50 | - | 0.50 | - | 0.50 | - | 0.50 | - | 0.50 | - | - | - | 0.50 | - | - | - | - | 0.50 | - |
| .....................................................................................................CATAACATACACACACACACGTA............................................................... | 23 | 2 | 122.50 | 122.50 | 9.00 | - | 46.00 | 13.50 | 2.50 | 10.00 | 3.50 | 3.00 | 0.50 | 4.00 | 2.00 | - | 3.00 | 10.00 | - | 4.50 | 1.50 | 0.50 | 0.50 | - | - | 1.50 | 1.00 | 0.50 | - | 0.50 | - | - | - | - | 0.50 | - | - | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTAT.............................................................. | 23 | 2 | 104.00 | 104.00 | 19.00 | 6.00 | - | - | 6.00 | - | 9.00 | 9.00 | 6.00 | 2.50 | 1.50 | 6.50 | 5.00 | 1.50 | - | 0.50 | 0.50 | 0.50 | 5.00 | - | 0.50 | 1.00 | 1.00 | - | 0.50 | 2.50 | 0.50 | - | 0.50 | - | 1.00 | - | 1.00 | 1.50 | - | - | - | 1.50 | 0.50 | - | 0.50 | 1.00 | - | 2.50 | 2.00 | 0.50 | - | - | 0.50 | - | - | 1.00 | 0.50 | - | 0.50 | - | 0.50 | 0.50 | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGT................................................................ | 23 | 2 | 99.00 | 99.00 | 8.00 | 11.00 | - | 2.00 | 2.50 | 7.00 | 4.50 | 2.00 | 0.50 | 1.00 | 2.00 | 0.50 | 0.50 | 0.50 | 3.00 | 1.50 | 3.00 | 3.50 | - | 3.50 | 3.50 | 0.50 | 1.50 | 2.00 | 0.50 | - | 2.50 | - | - | 0.50 | 2.00 | 3.50 | - | - | 1.50 | 2.50 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | 0.50 | 1.00 | 1.50 | 0.50 | - | 1.00 | - | - | 1.00 | - | 0.50 | - | 0.50 | 1.00 | 1.00 | 0.50 | - | - | 1.50 | - | - | - | 0.50 | - | - | 0.50 | 0.50 | - | 0.50 | - | - | - | - | 1.00 | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | 0.50 | - | - | - | - | 0.50 | - | 0.50 | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - |
| .....................................................................................................CATAACATACACACACACACGTAT.............................................................. | 24 | 2 | 81.00 | 81.00 | 10.50 | 4.00 | - | 2.00 | 3.50 | - | 5.00 | 6.50 | 1.50 | 12.50 | 3.50 | - | 5.00 | - | 0.50 | 1.00 | 0.50 | 0.50 | 1.50 | - | 0.50 | 0.50 | 1.00 | 2.00 | - | 2.00 | 0.50 | - | 4.50 | 0.50 | 1.00 | 0.50 | - | 1.00 | - | - | - | 0.50 | - | - | 1.00 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | 1.00 | - | 0.50 | - | - | 0.50 | - | 0.50 | - | - | - | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ......................................................................................................ATAACATACACACACACACGT................................................................ | 21 | 2 | 67.00 | 67.00 | 8.50 | 2.00 | - | 2.50 | 6.00 | - | 3.00 | 1.00 | 2.00 | 0.50 | 0.50 | 6.50 | 0.50 | 1.00 | 3.50 | 2.00 | 2.50 | - | 0.50 | 2.50 | 2.00 | 1.00 | 1.00 | 0.50 | 0.50 | - | - | 1.50 | - | 0.50 | - | 0.50 | 2.00 | - | - | 1.00 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | 0.50 | 0.50 | 0.50 | - | - | - | 1.00 | - | 0.50 | - | - | 1.00 | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | 0.50 | - | 0.50 | - | 0.50 | 1.00 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTA............................................................... | 22 | 2 | 63.00 | 63.00 | 6.00 | 1.00 | - | 15.50 | 7.50 | 1.50 | 3.50 | 2.00 | 1.50 | - | 1.50 | - | 2.50 | 5.00 | - | 3.00 | 0.50 | 1.00 | 1.00 | - | 0.50 | - | 1.50 | 0.50 | 0.50 | - | - | - | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | 1.00 | - | - | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | 0.50 | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACG................................................................. | 22 | 2 | 22.50 | 22.50 | - | 4.00 | - | - | - | - | 0.50 | 0.50 | 2.00 | - | 2.00 | - | 0.50 | - | 3.50 | 0.50 | - | - | - | - | - | 0.50 | 1.50 | - | - | - | - | - | - | - | 0.50 | - | 0.50 | 1.50 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | 0.50 | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACG................................................................. | 21 | 2 | 8.50 | 8.50 | - | 1.00 | - | - | - | - | 1.00 | 0.50 | 0.50 | - | 0.50 | 2.00 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGTA............................................................... | 24 | 2 | 8.00 | 8.00 | - | - | - | 1.50 | - | - | - | 1.00 | - | - | 0.50 | - | - | - | 0.50 | 0.50 | - | 0.50 | - | 0.50 | 0.50 | - | 0.50 | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGTAT.............................................................. | 25 | 2 | 7.50 | 7.50 | 1.50 | - | - | - | - | - | 0.50 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 1.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGTt............................................................... | 24 | T | 4.00 | 99.00 | - | - | - | - | - | 3.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTAa.............................................................. | 23 | A | 3.00 | 63.00 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACG................................................................. | 20 | 2 | 2.50 | 2.50 | - | - | - | - | - | - | - | 1.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TACATAACATACACACACACACGT................................................................ | 24 | 2 | 2.00 | 2.00 | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGa................................................................ | 23 | A | 2.00 | 22.50 | - | 0.50 | - | 0.50 | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TACATAACATACACACACACAt.................................................................. | 22 | T | 2.00 | 0.14 | - | 0.43 | - | - | - | - | - | - | - | - | - | 1.43 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAaat................................................................ | 23 | AAT | 1.50 | 0.00 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGTt............................................................... | 23 | T | 1.50 | 169.00 | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGTAa.............................................................. | 25 | A | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACtt................................................................ | 21 | TT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGca............................................................... | 23 | CA | 1.00 | 8.50 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTATt............................................................. | 24 | T | 1.00 | 104.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAaa................................................................. | 22 | AA | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGa................................................................ | 22 | A | 1.00 | 8.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAtgt................................................................ | 23 | TGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| .......................................................................................................TAACATACACACACACACGTAT.............................................................. | 22 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACtt................................................................ | 23 | TT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAtg................................................................. | 22 | TG | 1.00 | 0.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGc................................................................ | 23 | C | 1.00 | 22.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACAagta............................................................... | 23 | AGTA | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAagt................................................................ | 23 | AGT | 1.00 | 0.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGTATt............................................................. | 25 | T | 1.00 | 81.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ...................................................................................................TACATAACATACACACACACAC.................................................................. | 22 | 7 | 0.57 | 0.57 | - | - | - | 0.14 | - | - | - | - | - | - | 0.14 | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| .......................................................................................................TAACATACACACACACACGTAa.............................................................. | 22 | A | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGTga.............................................................. | 24 | GA | 0.50 | 169.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGTtt.............................................................. | 25 | TT | 0.50 | 99.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGTgt.............................................................. | 25 | GT | 0.50 | 99.00 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTATtt............................................................ | 25 | TT | 0.50 | 104.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAgat................................................................ | 23 | GAT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACatat.............................................................. | 25 | ATAT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGTAaa............................................................. | 25 | AA | 0.50 | 122.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAtata............................................................... | 24 | TATA | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACct................................................................ | 23 | CT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGTta.............................................................. | 24 | TA | 0.50 | 169.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACAtata............................................................... | 23 | TATA | 0.50 | 0.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGTATt............................................................. | 26 | T | 0.50 | 7.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACtt................................................................ | 22 | TT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAa.................................................................. | 21 | A | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTct.............................................................. | 23 | CT | 0.50 | 67.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAg.................................................................. | 21 | G | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTtt.............................................................. | 23 | TT | 0.50 | 67.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTAag............................................................. | 24 | AG | 0.50 | 63.00 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACAagt................................................................ | 22 | AGT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGc................................................................ | 21 | C | 0.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGTAa.............................................................. | 24 | A | 0.50 | 122.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGcat.............................................................. | 23 | CAT | 0.50 | 2.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AACATACACACACACACGTAT.............................................................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGga............................................................... | 23 | GA | 0.50 | 8.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TACATAACATACACACACACACG................................................................. | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACACGcat.............................................................. | 25 | CAT | 0.50 | 22.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACAtgt................................................................ | 22 | TGT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACtta............................................................... | 23 | TTA | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ACATAACATACACACACACAtgta............................................................... | 24 | TGTA | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACGTATc............................................................. | 24 | C | 0.50 | 104.00 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGg................................................................ | 22 | G | 0.50 | 8.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CATAACATACACACACACACGTc............................................................... | 23 | C | 0.50 | 169.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AACATACACACACACACGT................................................................ | 19 | 9 | 0.44 | 0.44 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAACATACACACACACACctat.............................................................. | 23 | CTAT | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TACATAACATACACACACACA................................................................... | 21 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAATTCATACTTATCCTCATGAGAGATCAGCCTGTGATTCCTCCATGTATGTGCATGTGTGTATAGTTGTGTGTGCATGTTCATGTCTATATTTGAATATACATAACATACACACACACACGTATAAACGCAAGCACACATACACAGACACAGGAATGGCATTCGCCAATCCCTTTGAATGATAAGT ..................................................(((((.((((((((((.(((((.((((.(((......))).))))))))).)))))))).)))))))................................................... ...........................................................60.................................................................127.......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR073954(GSM629280) total RNA. (blood) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR059768(GSM562830) Treg_control. (spleen) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR390298(GSM849856) cell line: NIH-3T3cell type: fibroblastinfect. (fibroblast) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR073955(GSM629281) total RNA. (blood) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | GSM510444(GSM510444) brain_rep5. (brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR037905(GSM510441) brain_rep2. (brain) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM314558(GSM314558) ESC wild type (454). (ESC) | GSM416732(GSM416732) MEF. (cell line) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | GSM261957(GSM261957) oocytesmallRNA-19to24. (oocyte) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR206942(GSM723283) other. (brain) | GSM475281(GSM475281) total RNA. (testes) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | GSM509280(GSM509280) small RNA cloning by length. (testes) |
|---|