| (3) B-CELL | (4) BRAIN | (2) CELL-LINE | (8) EMBRYO | (7) ESC | (3) LIVER | (3) LYMPH | (5) OTHER | (5) OTHER.mut | (1) PIWI.mut | (3) SPLEEN | (13) TESTES | (2) THYMUS | (1) TOTAL-RNA |
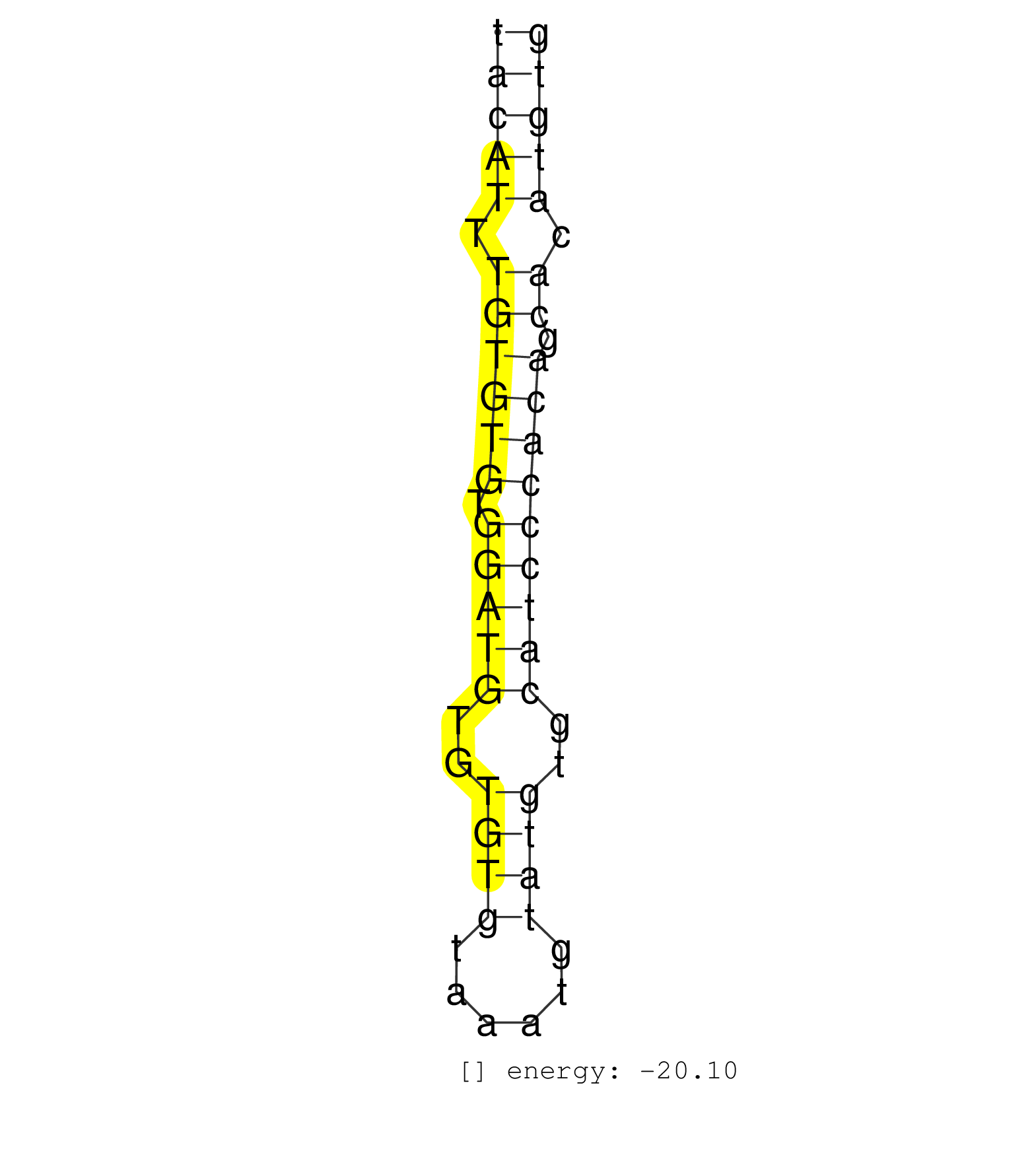
| GCAAAGGGCACTCGGGCCTTTCACTCTGTGCAGACCACTCTGCAGATCTTTACATTTGTGTGTGGATGTGTGTGTAAATGTATGTGCATCCCACAGCACATGTGCAGAGGGCATTGGCTTACTCCTTCTACCACGTGGAGCCTGAAGACTGAAC ..................................................(((((.((((((.(((((..((((......))))..))))))))).)).))))).................................................. ..................................................51...................................................104................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | mjLiverWT2() Liver Data. (liver) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM361408(GSM361408) WholeCerebellum_P6_wt_rep1. (brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR059765(GSM562827) DN3_control. (thymus) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | GSM510456(GSM510456) newborn_rep12. (total RNA) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................ATTTGTGTGTGGATGTGTGTat............................................................................... | 22 | AT | 34.00 | 9.00 | 1.00 | 5.67 | 7.33 | 0.67 | - | 1.67 | 0.33 | 0.33 | 0.67 | - | 1.00 | 1.33 | 0.33 | 0.33 | 0.33 | 0.33 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 0.33 | 0.67 | 0.33 | 0.67 | 0.33 | 0.33 | 0.67 | 0.67 | 0.33 | 0.33 | 0.67 | - | - | - | 0.33 | 0.33 | - | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | 0.33 | - | 0.33 | 0.33 | 0.33 | 0.33 | - |
| .....................................................ATTTGTGTGTGGATGTGTGT................................................................................. | 20 | 3 | 9.00 | 9.00 | 0.67 | 2.33 | - | - | - | - | 0.33 | - | 0.33 | 1.00 | - | - | - | 0.67 | 0.67 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | 0.67 | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | 0.33 |
| ................................................................GATGTGTGTGTAAATGTAcat..................................................................... | 21 | CAT | 5.00 | 0.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATTTGTGTGTGGATGTGTGTa................................................................................ | 21 | A | 4.00 | 9.00 | 0.33 | 0.67 | - | 0.33 | - | - | 0.67 | - | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GATGTGTGTGTAAATGTAcatg.................................................................... | 22 | CATG | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................ACATTTGTGTGTGGATGTGTGTat............................................................................... | 24 | AT | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................ATGTGTGTGTAAATGTAcat..................................................................... | 20 | CAT | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GATGTGTGTGTAAATGTATat..................................................................... | 21 | AT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GGATGTGTGTGTAAAaacc........................................................................ | 19 | AACC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TTTGTGTGTGGATGTGTGTaa............................................................................... | 21 | AA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................ATGTGTGTGTAAATGTAcag..................................................................... | 20 | CAG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GATGTGTGTGTAAATGTAcag..................................................................... | 21 | CAG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATTTGTGTGTGGATGTGTGTaa............................................................................... | 22 | AA | 0.67 | 9.00 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATTTGTGTGTGGATGTGTG.................................................................................. | 19 | 5 | 0.40 | 0.40 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATTTGTGTGTGGATGTGTGTag............................................................................... | 22 | AG | 0.33 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATTTGTGTGTGGATGTGTGTatt.............................................................................. | 23 | ATT | 0.33 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATTTGTGTGTGGATGTGTGTc................................................................................ | 21 | C | 0.33 | 9.00 | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATTTGTGTGTGGATGTGTGTata.............................................................................. | 23 | ATA | 0.33 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCAAAGGGCACTCGGGCCTTTCACTCTGTGCAGACCACTCTGCAGATCTTTACATTTGTGTGTGGATGTGTGTGTAAATGTATGTGCATCCCACAGCACATGTGCAGAGGGCATTGGCTTACTCCTTCTACCACGTGGAGCCTGAAGACTGAAC ..................................................(((((.((((((.(((((..((((......))))..))))))))).)).))))).................................................. ..................................................51...................................................104................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | mjLiverWT2() Liver Data. (liver) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM361408(GSM361408) WholeCerebellum_P6_wt_rep1. (brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR059765(GSM562827) DN3_control. (thymus) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | GSM510456(GSM510456) newborn_rep12. (total RNA) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................ataTGCATCCCACAGCAC....................................................... | 18 | ata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................atgTTTGTGTGTGGATGTGTGT................................................................................. | 22 | atg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........tgtgGGGCCTTTCACTCTG.............................................................................................................................. | 19 | tgtg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................gcaTACTCCTTCTACCAC.................... | 18 | gca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATTTGTGTGTGGATGTGTGTGT............................................................................... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |