| (4) AGO2.ip | (2) B-CELL | (18) BRAIN | (8) CELL-LINE | (1) DGCR8.mut | (17) EMBRYO | (13) ESC | (5) FIBROBLAST | (2) LIVER | (1) LYMPH | (3) OTHER | (2) OTHER.mut | (3) PIWI.ip | (3) SPLEEN | (11) TESTES | (3) THYMUS | (2) TOTAL-RNA | (2) UTERUS |
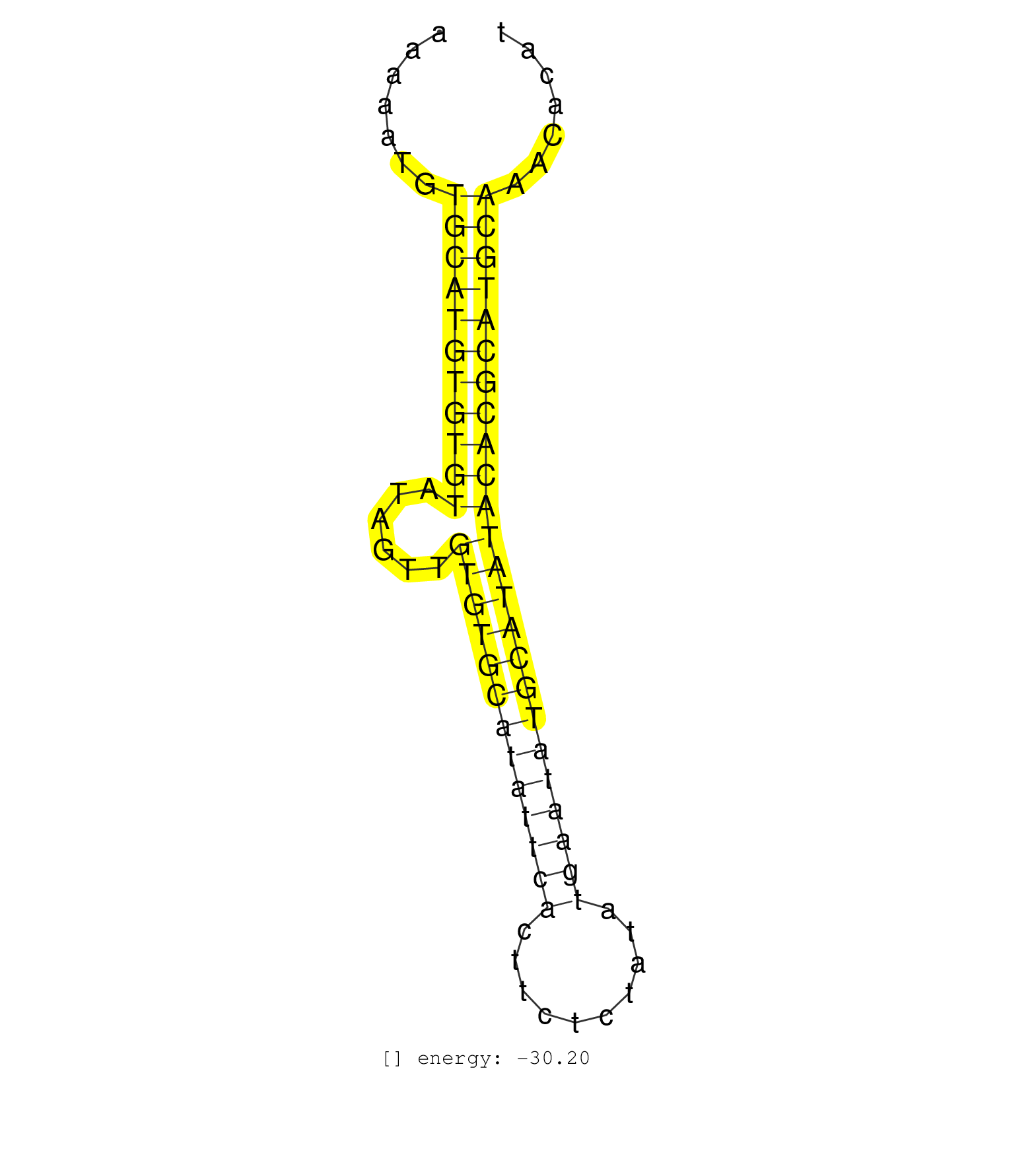
| TCAAATTTGTGCTGAATTTCATGATGAGTTCATGACCAGCATGGGATTCATCTTATGCCCACCAAAAATGTGCATGTGTGTATAGTTGTGTGCATATTCACTTCTCTATATGAATATGCATATACACGCATGCAAACACATTCATAGGAATGGCCAGAGCTATATCTCAGTTAGGAGGTGAGCACTCTTGCCAGAGCTAGGAAGATGAGGAGTTCAAATTCATGTTT .........................................................(((((((((((......(((((((((((((..........))))))))))))))))))))))))......................................................... ...............................................................64...........................................................................141.................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM475281(GSM475281) total RNA. (testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR037923(GSM510461) e9p5_rep1. (embryo) | mjTestesWT4() Testes Data. (testes) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR037905(GSM510441) brain_rep2. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR029123(GSM416611) NIH3T3. (cell line) | GSM416732(GSM416732) MEF. (cell line) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037937(GSM510475) 293cand2. (cell line) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR206941(GSM723282) other. (brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR206942(GSM723283) other. (brain) | SRR037919(GSM510457) e12p5_rep1. (embryo) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR037906(GSM510442) brain_rep3. (brain) | SRR073954(GSM629280) total RNA. (blood) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR037920(GSM510458) e12p5_rep2. (embryo) | GSM361407(GSM361407) CGNP_P6_Ptc+-_Ink4c--_rep5. (brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR059769(GSM562831) Treg_Drosha. (spleen) | GSM361415(GSM361415) WholeCerebellum_P6_p53--_Ink4c--_rep4. (brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR059771(GSM562833) CD4_control. (spleen) | GSM261958(GSM261958) oocytesmallRNA-24to30. (oocyte) | GSM510444(GSM510444) brain_rep5. (brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR037904(GSM510440) brain_rep1. (brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR206939(GSM723280) other. (brain) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | GSM314559(GSM314559) ESC dgcr8 (454). (ESC) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037930(GSM510468) e7p5_rep4. (embryo) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................TGCATATACACGCATGCAAACA......................................................................................... | 22 | 2 | 38.00 | 38.00 | 7.50 | 2.50 | - | - | 4.00 | 6.00 | 3.00 | 2.50 | 0.50 | 1.00 | 3.00 | - | 3.50 | - | - | - | - | - | 0.50 | - | - | - | 1.50 | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAAC.......................................................................................... | 21 | 2 | 30.00 | 30.00 | 4.00 | 3.50 | 6.50 | 2.00 | 0.50 | 1.00 | 1.50 | 1.00 | 1.50 | 2.00 | - | - | - | - | - | - | - | - | 0.50 | 1.50 | - | 0.50 | 1.00 | 0.50 | - | - | - | - | 0.50 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTGCATGTGTGTATAGTTGTGTGC...................................................................................................................................... | 25 | 9 | 15.33 | 15.33 | 1.33 | - | - | 0.33 | - | 0.11 | 0.11 | - | 0.11 | - | - | - | - | 2.78 | 0.22 | 0.33 | 0.56 | 0.33 | 0.22 | - | - | 0.22 | - | 1.33 | 0.33 | 0.56 | 1.56 | 0.22 | 0.11 | 0.33 | 0.78 | 0.11 | - | - | - | - | - | - | - | 0.78 | 0.11 | 0.11 | 0.22 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | 0.22 | - | 0.33 | - | - | - | 0.33 | - | - | 0.22 | 0.22 | 0.22 | - | 0.11 | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | 0.11 | 0.11 | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACAC........................................................................................ | 23 | 2 | 12.00 | 12.00 | 2.00 | 3.50 | - | 1.50 | 2.00 | 1.00 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAA........................................................................................... | 20 | 3 | 10.00 | 10.00 | 1.33 | - | - | 1.33 | - | - | 0.33 | 0.33 | 1.33 | 0.33 | - | - | 0.33 | - | 0.33 | - | - | - | - | 1.00 | 2.00 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCAA............................................................................................ | 21 | 2 | 9.00 | 9.00 | 1.50 | - | - | 0.50 | 0.50 | - | - | 0.50 | - | - | 1.50 | - | - | - | - | 1.00 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCAAAC.......................................................................................... | 23 | 2 | 8.50 | 8.50 | 5.50 | 0.50 | - | - | - | - | 1.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACt......................................................................................... | 22 | T | 7.00 | 30.00 | 2.00 | - | 0.50 | 1.00 | - | 0.50 | 1.50 | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATGCATATACACGCATGCAAAC.......................................................................................... | 22 | 2 | 6.50 | 6.50 | 0.50 | 2.00 | - | 1.50 | - | - | - | - | 0.50 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GTGCATGTGTGTATAGTTGTGTGC...................................................................................................................................... | 24 | 9 | 5.33 | 5.33 | 1.11 | - | 0.44 | 0.11 | - | 0.11 | - | 0.11 | - | 0.11 | - | - | - | 0.44 | - | 0.11 | 0.11 | 0.22 | 0.11 | 0.11 | - | - | - | 0.11 | 0.33 | - | - | 0.11 | - | - | 0.22 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | 0.11 | 0.11 | - | - | - | - | 0.22 | - | - | 0.22 | - | - | - | - | - | - | - | - | 0.11 | 0.11 | - | - | 0.11 | 0.11 | - | 0.11 |
| ..................................................................................................................TATGCATATACACGCATGCA............................................................................................. | 20 | 7 | 5.29 | 5.29 | 0.29 | - | 0.14 | 0.71 | - | 0.14 | - | 0.14 | 0.29 | - | - | 0.43 | 0.14 | - | - | - | - | - | - | 0.14 | 0.86 | - | - | - | - | 0.71 | - | 0.14 | - | 0.14 | - | - | 0.14 | - | - | - | - | 0.14 | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCAAt........................................................................................... | 22 | T | 5.00 | 9.00 | 1.50 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAcact......................................................................................... | 22 | CACT | 5.00 | 0.00 | 1.00 | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTGCATGTGTGTATAGTTGTGTGCATA................................................................................................................................... | 28 | 9 | 4.56 | 4.56 | - | - | - | - | - | - | - | - | - | 0.44 | - | - | - | - | - | 0.78 | 1.00 | 0.67 | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.56 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | 0.22 | - | - | - | - | - | 0.11 | - | - | 0.11 | - | - | - | - | - | - | - | - |
| .................................................................................................................ATATGCATATACACGCATGCAA............................................................................................ | 22 | 2 | 3.50 | 3.50 | - | - | - | 0.50 | - | - | 0.50 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAttat......................................................................................... | 22 | TTAT | 3.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATGCATATACACGCATGCAAA........................................................................................... | 21 | 2 | 3.00 | 3.00 | - | - | 1.00 | - | 1.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTGCATGTGTGTATAGTTGTGTGCAT.................................................................................................................................... | 27 | 9 | 2.56 | 2.56 | 0.78 | - | - | 0.11 | - | - | - | - | 0.11 | - | - | - | - | - | 0.11 | 0.33 | - | 0.44 | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | 0.33 | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAA............................................................................................ | 19 | 4 | 2.50 | 2.50 | 0.75 | - | - | - | - | 0.75 | - | 0.75 | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACACA....................................................................................... | 24 | 2 | 2.50 | 2.50 | - | 1.50 | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACc......................................................................................... | 22 | C | 2.50 | 30.00 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACACt....................................................................................... | 24 | T | 2.50 | 12.00 | - | 1.00 | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAtttt......................................................................................... | 22 | TTTT | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATGCATATACACGCATGCAcact......................................................................................... | 23 | CACT | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TTCACTTCTCTATATGcata............................................................................................................... | 20 | CATA | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATATGCATATACACGCATGCA............................................................................................. | 21 | 7 | 2.00 | 2.00 | 0.43 | - | - | 0.14 | 0.29 | - | - | 0.14 | - | - | - | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | 0.43 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GTGCATGTGTGTATAGTTGTGTGCAT.................................................................................................................................... | 26 | 9 | 1.89 | 1.89 | 0.56 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.44 | 0.33 | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................ATGTGCATGTGTGTATAGTTGTGTGC...................................................................................................................................... | 26 | 9 | 1.67 | 1.67 | 0.44 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCAAA........................................................................................... | 22 | 2 | 1.50 | 1.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACAt........................................................................................ | 23 | T | 1.50 | 38.00 | - | 0.50 | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAg............................................................................................ | 19 | G | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGtaat........................................................................................... | 20 | TAAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CACCAAAAATGTGCATagcc.................................................................................................................................................... | 20 | AGCC | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCActt.......................................................................................... | 21 | CTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TATACACGCATGCAAACAC........................................................................................ | 19 | 2 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACta........................................................................................ | 23 | TA | 1.00 | 30.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATGCATATACACGCATGCAAga.......................................................................................... | 22 | GA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAga........................................................................................... | 20 | GA | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACAaa....................................................................................... | 24 | AA | 1.00 | 38.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGTGTGTATAGTTGTGTGCATt................................................................................................................................... | 22 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAAt.......................................................................................... | 21 | T | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATATGCATATACACGCATGCAAt........................................................................................... | 23 | T | 1.00 | 3.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAcgct......................................................................................... | 22 | CGCT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................AGAGCTAGGAAGATGtgtt................ | 19 | TGTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTGCATGTGTGTATAGTTGTGTGCA..................................................................................................................................... | 26 | 9 | 1.00 | 1.00 | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | 0.22 | 0.11 | - | - | 0.33 | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGC.............................................................................................. | 19 | 7 | 0.86 | 0.86 | 0.14 | - | - | 0.14 | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GTGCATGTGTGTATAGTTGTGTGCATA................................................................................................................................... | 27 | 9 | 0.78 | 0.78 | 0.22 | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GTGCATGTGTGTATAGTTGTGTGCA..................................................................................................................................... | 25 | 9 | 0.56 | 0.56 | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | 0.22 | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTGCATGTGTGTATAGTTGTGTGCATt................................................................................................................................... | 28 | T | 0.56 | 2.56 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCA................................................................................................. | 16 | 10 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGCATGTGTGTATAGTTGTGTGCATATTatt.............................................................................................................................. | 31 | ATT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCAAtt.......................................................................................... | 23 | TT | 0.50 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACg......................................................................................... | 22 | G | 0.50 | 30.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TATATGAATATGCATATACACGCATGCAAA........................................................................................... | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GCATGTGTGTATAGTTGTGTGCAa.................................................................................................................................... | 24 | A | 0.50 | 0.00 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAttt......................................................................................... | 22 | TTT | 0.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCAAAa.......................................................................................... | 23 | A | 0.50 | 1.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATATGCATATACACGCATGCAAA........................................................................................... | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGCATGTGTGTATAGTTGTGTGCATt................................................................................................................................... | 26 | T | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACACAa...................................................................................... | 25 | A | 0.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GCATGTGTGTATAGTTGTGTGCATATa................................................................................................................................. | 27 | A | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCAAAt.......................................................................................... | 23 | T | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAACAa........................................................................................ | 23 | A | 0.50 | 38.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATGCATATACACGCATGCAAACA......................................................................................... | 23 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................ATGCATATACACGCATGCAAACt......................................................................................... | 23 | T | 0.50 | 6.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................ATGTGCATGTGTGTATAGTTGTGTGCATA................................................................................................................................... | 29 | 9 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TATACACGCATGCAAAC.......................................................................................... | 17 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATATGCATATACACGCAT................................................................................................ | 18 | 7 | 0.29 | 0.29 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................GCATATACACGCATGCAA............................................................................................ | 18 | 4 | 0.25 | 0.25 | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAttat........................................................................................ | 23 | TTAT | 0.25 | 2.50 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAt........................................................................................... | 20 | T | 0.25 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCAT................................................................................................ | 17 | 9 | 0.22 | 0.22 | - | - | - | - | 0.11 | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATATGCATATACACGCATGC.............................................................................................. | 20 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGg.............................................................................................. | 19 | G | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGa.............................................................................................. | 19 | A | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCAcaa.......................................................................................... | 23 | CAA | 0.14 | 5.29 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATGCga............................................................................................ | 21 | GA | 0.14 | 0.86 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCATG............................................................................................... | 18 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATATGCATATACACGCATGCcat........................................................................................... | 23 | CAT | 0.14 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................ATACACGCATGCAAAC.......................................................................................... | 16 | 8 | 0.12 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTGCATGTGTGTATAGTTGTGTGCtt.................................................................................................................................... | 27 | TT | 0.11 | 15.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AATGTGCATGTGTGTATAGTTGTGTGCATA................................................................................................................................... | 30 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCAcgca............................................................................................. | 20 | CGCA | 0.10 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TATGCATATACACGCAggca............................................................................................. | 20 | GGCA | 0.10 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCAAATTTGTGCTGAATTTCATGATGAGTTCATGACCAGCATGGGATTCATCTTATGCCCACCAAAAATGTGCATGTGTGTATAGTTGTGTGCATATTCACTTCTCTATATGAATATGCATATACACGCATGCAAACACATTCATAGGAATGGCCAGAGCTATATCTCAGTTAGGAGGTGAGCACTCTTGCCAGAGCTAGGAAGATGAGGAGTTCAAATTCATGTTT .........................................................(((((((((((......(((((((((((((..........))))))))))))))))))))))))......................................................... ...............................................................64...........................................................................141.................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM475281(GSM475281) total RNA. (testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR037923(GSM510461) e9p5_rep1. (embryo) | mjTestesWT4() Testes Data. (testes) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR037905(GSM510441) brain_rep2. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR029123(GSM416611) NIH3T3. (cell line) | GSM416732(GSM416732) MEF. (cell line) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037937(GSM510475) 293cand2. (cell line) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR206941(GSM723282) other. (brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR206942(GSM723283) other. (brain) | SRR037919(GSM510457) e12p5_rep1. (embryo) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR037906(GSM510442) brain_rep3. (brain) | SRR073954(GSM629280) total RNA. (blood) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR037920(GSM510458) e12p5_rep2. (embryo) | GSM361407(GSM361407) CGNP_P6_Ptc+-_Ink4c--_rep5. (brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR059769(GSM562831) Treg_Drosha. (spleen) | GSM361415(GSM361415) WholeCerebellum_P6_p53--_Ink4c--_rep4. (brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR059771(GSM562833) CD4_control. (spleen) | GSM261958(GSM261958) oocytesmallRNA-24to30. (oocyte) | GSM510444(GSM510444) brain_rep5. (brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR037904(GSM510440) brain_rep1. (brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR206939(GSM723280) other. (brain) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | GSM314559(GSM314559) ESC dgcr8 (454). (ESC) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037930(GSM510468) e7p5_rep4. (embryo) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................tcCCCACCAAAAATGTGC.......................................................................................................................................................... | 18 | tc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCATATACACGCATGCAAA........................................................................................... | 20 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTGCATGTGTGTATAGTTGTGTGC...................................................................................................................................... | 25 | 9 | 0.22 | 0.22 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | 0.11 | - |