| (1) BRAIN | (1) CELL-LINE | (4) EMBRYO | (1) ESC | (1) HEART | (1) LIVER | (2) OTHER.mut | (2) PIWI.ip | (2) PIWI.mut | (8) TESTES |
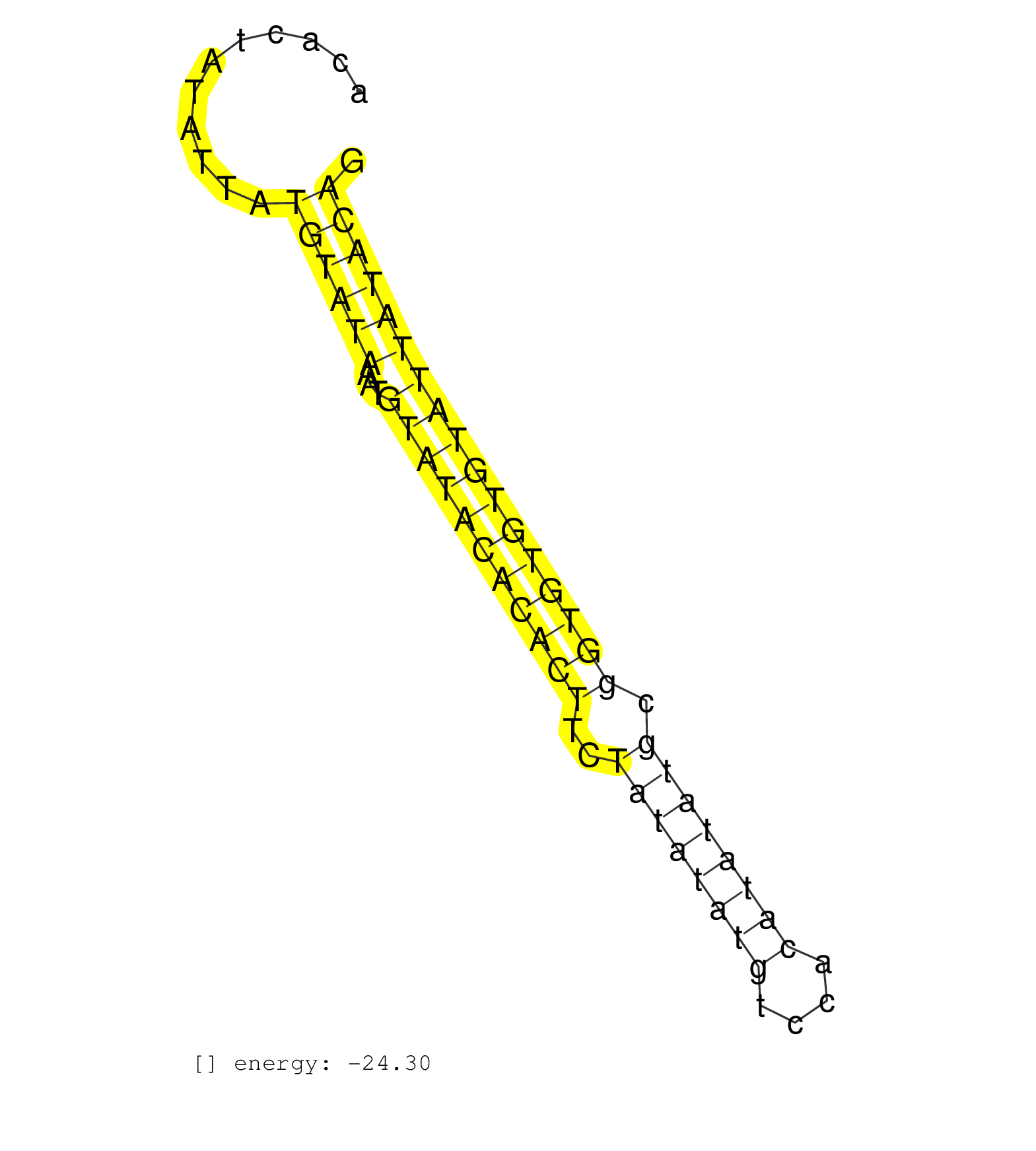
| ACGTGCTAGTGTCTGAACCCAGTTCAGCATATCTCCAGTTGAAACAGTATACACTATATTATGTATAAATGTATACACACTTCTATATATGTCCACATATATGCGGTGTGTGTATTATACAGGTATAGGTGTGTGTGCACGCACACAGGTGCACATAGCATATCAAGTGTTCATTACAAATCC .............................................................((((((...(((((((((((..((((((((....)))))))).)))))))))))))))))................................................... ..................................................51.....................................................................122........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037936(GSM510474) 293cand1. (cell line) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR095855BC2(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................GGTATAGGTGTGTGTGCACGCAC....................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CGTGCTAGTGTCTGAACCCA.................................................................................................................................................................. | 20 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TATCTCCAGTTGAAACAGTATACACTATAT............................................................................................................................ | 30 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................TAGGTGTGTGTGCACGCACACAGGT................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TATCTCCAGTTGAAACAGTATACACTAT.............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................ATACACTATATTATGTATAAAT................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................TTATACAGGTATAGGTGTGTGTGCA............................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................GCATATCTCCAGTTGAAACAG........................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................TATTATGTATAAATGTATACACACTTC.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................ATCTCCAGTTGAAACAGTAT..................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TAGGTGTGTGTGCACGCACACAGGTGt............................... | 27 | T | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ATATTATGTATAAATGTATACACACTTCT................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TCTCCAGTTGAAACAGTATACACTATAT............................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............GAACCCAGTTCAGCATATCTCCAGTT............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATTATACAGGTATAGGT..................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................GTGTGTGTATTATACAGG............................................................ | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TATCTCCAGTTGAAACAGTATACACTATATTA.......................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ATGCGGTGTGTGTATTAT................................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................AGCATATCAAGTGTTCATT........ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................TGTGCACGCACACAGGTGCAC............................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ACGTGCTAGTGTCTGAACCCAGTTCAGCATATCTCCAGTTGAAACAGTATACACTATATTATGTATAAATGTATACACACTTCTATATATGTCCACATATATGCGGTGTGTGTATTATACAGGTATAGGTGTGTGTGCACGCACACAGGTGCACATAGCATATCAAGTGTTCATTACAAATCC .............................................................((((((...(((((((((((..((((((((....)))))))).)))))))))))))))))................................................... ..................................................51.....................................................................122........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037936(GSM510474) 293cand1. (cell line) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR095855BC2(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................ATGTATAAATGTATACACACTTCT................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TCTGAACCCAGTTCAGCATA........................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TCTGAACCCAGTTCA............................................................................................................................................................. | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |