| (4) AGO2.ip | (11) BRAIN | (5) EMBRYO | (3) ESC | (3) FIBROBLAST | (3) LIVER | (4) OTHER | (2) SPLEEN | (3) TESTES | (2) THYMUS |
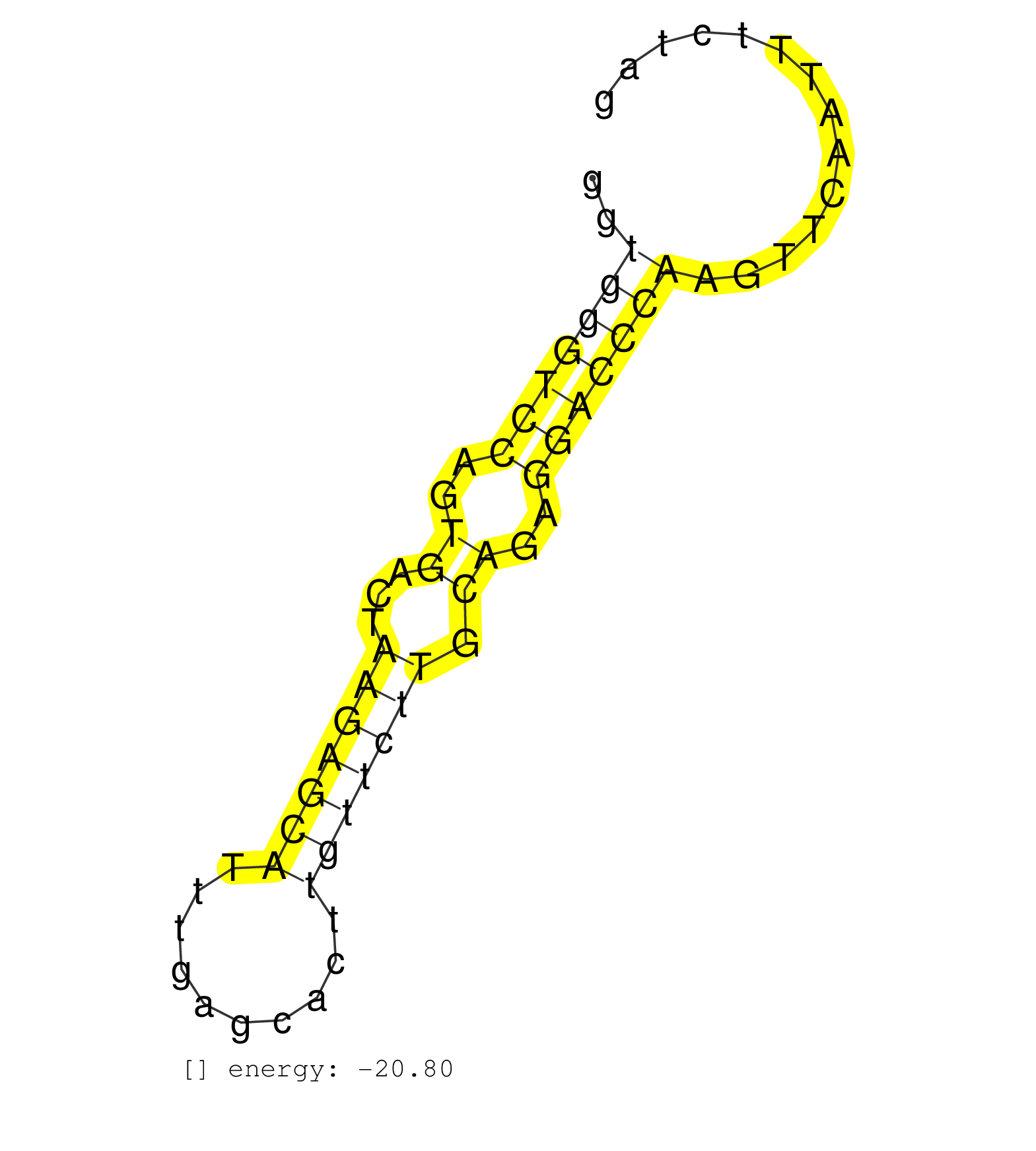
| TTAATTATTTTTTTAATTGTTATTTTCCTATGTCAGAAATTGATGGGGACTAGAAAGGTGGGTCCAGTGACTAAGAGCATTTGAGCACTTGTTCTTGCAGAGGACCCAAGTTCAATTTCTAGCACCTATATGGTGGCTCACAGCAATCTGTAACTCTAGCTCCAGGGGATTG ..........................................................(((((((..((...(((((((..........))))))).))..)))))))................................................................ ........................................................57...............................................................122................................................ | Size | Perfect hit | Total Norm | Perfect Norm | mjLiverWT1() Liver Data. (liver) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR059768(GSM562830) Treg_control. (spleen) | GSM261957(GSM261957) oocytesmallRNA-19to24. (oocyte) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | GSM261959(GSM261959) oocytesmallRNA-19to30. (oocyte) | SRR077865(GSM637802) 18-30 nt small RNAs. (liver) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR059765(GSM562827) DN3_control. (thymus) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | GSM361408(GSM361408) WholeCerebellum_P6_wt_rep1. (brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM361430(GSM361430) WholeCerebellum_1month_Ptc+-_Ink4c--_rep2. (brain) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................AGTGACTAAGAGCATgtgg........................................................................................ | 19 | GTGG | 16.00 | 0.00 | 7.00 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AGTGACTAAGAGCATgtg......................................................................................... | 18 | GTG | 12.00 | 0.00 | - | 3.00 | - | 2.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| .............................................................GTCCAGTGACTAAGAGCAT............................................................................................ | 19 | 1 | 4.00 | 4.00 | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTCCAGTGACTAAGAGCATgtgg........................................................................................ | 23 | GTGG | 3.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGGTCCAGTGACTAAGAGCATg........................................................................................... | 22 | G | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AGTGACTAAGAGCATat.......................................................................................... | 17 | AT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGACTAAGAGCATTTGgtgg..................................................................................... | 20 | GTGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGCAGAGGACCCAAGTagaa......................................................... | 20 | AGAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTCCAGTGACTAAGAGCg............................................................................................. | 18 | G | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GTCCAGTGACTAAGAGC.............................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................TTGCAGAGGACCCAAGTTCAAaga...................................................... | 24 | AGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................AGTGACTAAGAGCATatg......................................................................................... | 18 | ATG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGTCCAGTGACTAAGAGCA............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................AAATTGATGGGGACTct....................................................................................................................... | 17 | CT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTGCAGAGGACCCAAGTTCAAaa....................................................... | 23 | AA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGGTGGCTCACAGCAATCTGTAAtgga............... | 27 | TGGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGCACCTATATGGTGtgac................................. | 19 | TGAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................GTCCAGTGACTAAGAGCA............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................ATATGGTGGCTCACAtatg.......................... | 19 | TATG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................TGAGCACTTGTTCTTGCAGAGGA.................................................................... | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| TTAATTATTTTTTTAATTGTTATTTTCCTATGTCAGAAATTGATGGGGACTAGAAAGGTGGGTCCAGTGACTAAGAGCATTTGAGCACTTGTTCTTGCAGAGGACCCAAGTTCAATTTCTAGCACCTATATGGTGGCTCACAGCAATCTGTAACTCTAGCTCCAGGGGATTG ..........................................................(((((((..((...(((((((..........))))))).))..)))))))................................................................ ........................................................57...............................................................122................................................ | Size | Perfect hit | Total Norm | Perfect Norm | mjLiverWT1() Liver Data. (liver) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR059768(GSM562830) Treg_control. (spleen) | GSM261957(GSM261957) oocytesmallRNA-19to24. (oocyte) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | GSM261959(GSM261959) oocytesmallRNA-19to30. (oocyte) | SRR077865(GSM637802) 18-30 nt small RNAs. (liver) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR059765(GSM562827) DN3_control. (thymus) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | GSM361408(GSM361408) WholeCerebellum_P6_wt_rep1. (brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM361430(GSM361430) WholeCerebellum_1month_Ptc+-_Ink4c--_rep2. (brain) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................gtcaAGTGACTAAGAGCAT............................................................................................ | 19 | gtca | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................tcaAGTGACTAAGAGCAT............................................................................................ | 18 | tca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................CCAAGTTCAATTTCTAGCA................................................ | 19 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |