| (6) AGO2.ip | (1) AGO3.ip | (1) B-CELL | (13) BRAIN | (5) EMBRYO | (3) ESC | (1) FIBROBLAST | (1) LIVER | (1) LUNG | (2) OTHER | (5) OTHER.mut | (1) PIWI.ip | (1) SKIN | (1) SPLEEN | (11) TESTES | (1) THYMUS |
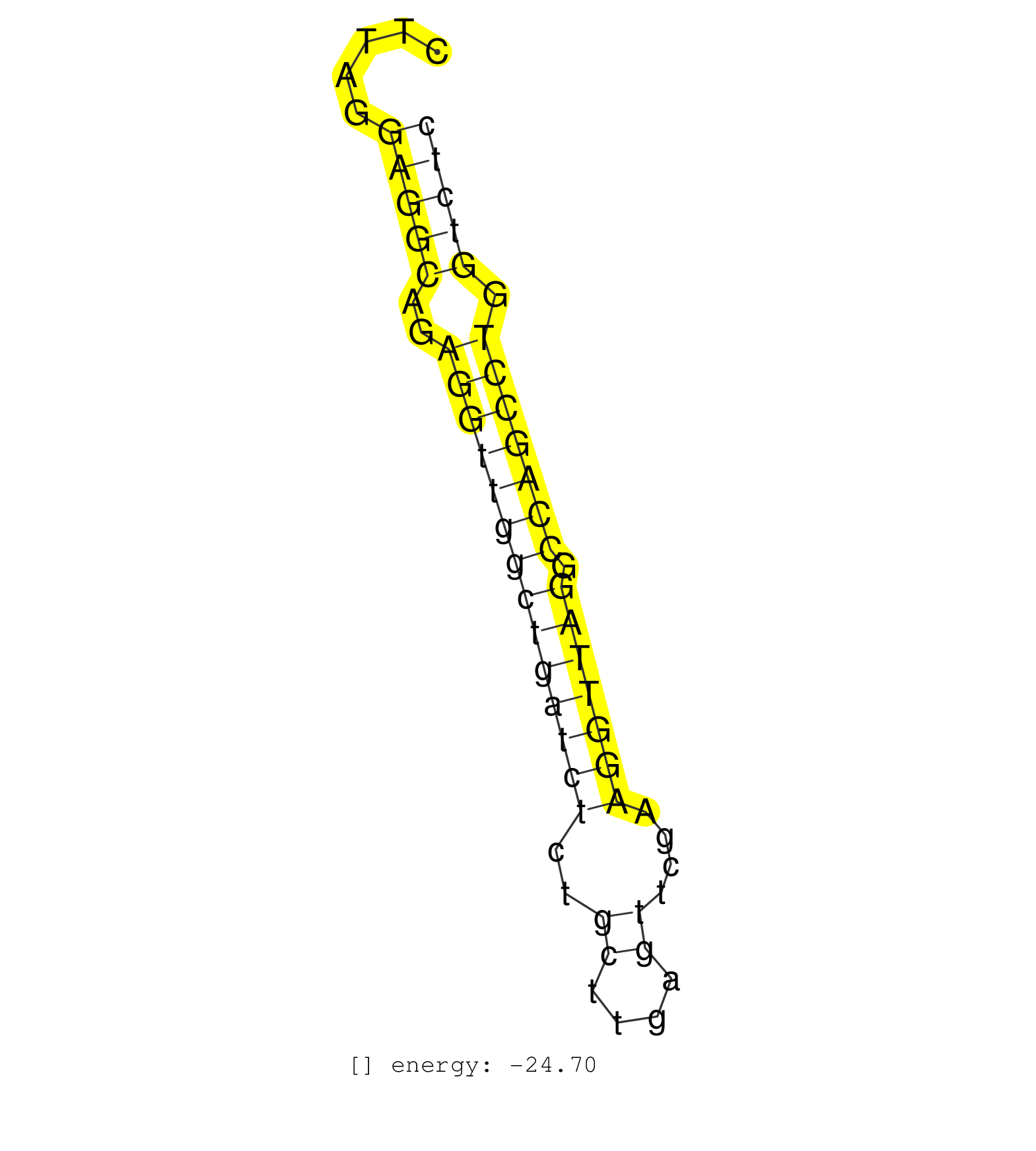
| TTAAAGAAAGCACTACTTGGTGACACTGACACATGCATTTAATCCTAGTACTTAGGAGGCAGAGGTTGGCTGATCTCTGCTTGAGTTCGAAGGTTAGGCCAGCCTGGTCTCCAAAGAGAGTTCCAAGACAGCCAGGACTACGCAGAGAAACCCTGTCTCAAAGAAAC .......................................................(((((..((((((((((((((..((....))....))))))).))))))).)))))........................................................ ..................................................51..........................................................111...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | mjTestesWT3() Testes Data. (testes) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR073954(GSM629280) total RNA. (blood) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................GTTAGGCCAGCCTGGcgtt........................................................ | 19 | CGTT | 4.00 | 0.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGGTTAGGCCAGCCTGGcgtt........................................................ | 22 | CGTT | 2.33 | 0.33 | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | 0.33 | - | 0.33 | 0.33 | - | - | - |
| .........................................................GGCAGAGGTTGGCTGttct........................................................................................... | 19 | TTCT | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACGCAGAGAAACCCTGTCTtt....... | 21 | TT | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GGCAGAGGTTGGCTGtttt........................................................................................... | 19 | TTTT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGGAGGCAGAGGTTGcag................................................................................................ | 18 | CAG | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GAAGGTTAGGCCAGCCT.............................................................. | 17 | 2 | 1.00 | 1.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CAAGACAGCCAGGACTACaata...................... | 22 | AATA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTACGCAGAGAAACCCTGTCTtttt..... | 25 | TTTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AGAGAGTTCCAAGACAGCCAGGgcca........................... | 26 | GCCA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GGCAGAGGTTGGCTGttcc........................................................................................... | 19 | TTCC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GGAGGCAGAGGTTGGCTGtg............................................................................................. | 20 | TG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGAGTTCGAAGGTTAtat.................................................................... | 18 | TAT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CAAGACAGCCAGGACTACGCAGAGAA.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GGCAGAGGTTGGCTGATtt........................................................................................... | 19 | TT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGGTTAGGCCAGCCTGGcgt......................................................... | 21 | CGT | 0.67 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | 0.33 | - | - |
| ........................................................................................GAAGGTTAGGCCAGCCTGG............................................................ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ........................................................................................GAAGGTTAGGCCAGCCTGGc........................................................... | 20 | C | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GAAGGTTAGGCCAGCCTGGcg.......................................................... | 21 | CG | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................GAGTTCCAAGACAGCCcgca.............................. | 20 | CGCA | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGGTTAGGCCAGCCTGG............................................................ | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| .........................................................................................AAGGTTAGGCCAGCCTGGcgg......................................................... | 21 | CGG | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - |
| TTAAAGAAAGCACTACTTGGTGACACTGACACATGCATTTAATCCTAGTACTTAGGAGGCAGAGGTTGGCTGATCTCTGCTTGAGTTCGAAGGTTAGGCCAGCCTGGTCTCCAAAGAGAGTTCCAAGACAGCCAGGACTACGCAGAGAAACCCTGTCTCAAAGAAAC .......................................................(((((..((((((((((((((..((....))....))))))).))))))).)))))........................................................ ..................................................51..........................................................111...................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | mjTestesWT3() Testes Data. (testes) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR073954(GSM629280) total RNA. (blood) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................tGCCAGCCTGGTCTCCAAA.................................................... | 19 | t | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................agttCAGCCTGGTCTCCAA..................................................... | 19 | agtt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ttACTACGCAGAGAAACCCTG............ | 21 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................agcGACACTGACACATGCA.................................................................................................................................. | 19 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................agaaGCCAGCCTGGTCTCCAA..................................................... | 21 | agaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................ttgCACATGCATTTAATCCTA........................................................................................................................ | 21 | ttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................tcctGGACTACGCAGAGAA.................. | 19 | tcct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ttttACCCTGTCTCAAAGA... | 19 | tttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................tAGTTCCAAGACAGCCAGGACTACG......................... | 25 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ttTTAGGCCAGCCTGGTCTCC....................................................... | 21 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................tcaCAAGACAGCCAGGAC............................. | 18 | tca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................cggCAGCCTGGTCTCCAAAGAGAG............................................... | 24 | cgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CCAGGACTACGCAGAGAAACCCTGTCTCA....... | 29 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| .......................................................................................................................................GACTACGCAGAGAAACCCTGTCTCAA...... | 26 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |
| .....................................................................................................................................AGGACTACGCAGAGAAACCCTGTCTCAA...... | 28 | 10 | 0.10 | 0.10 | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |