| (2) B-CELL | (1) BRAIN | (6) CELL-LINE | (13) EMBRYO | (12) ESC | (3) FIBROBLAST | (1) LIVER | (1) OTHER | (4) OTHER.mut | (1) PIWI.ip | (4) SPLEEN | (14) TESTES | (4) THYMUS |
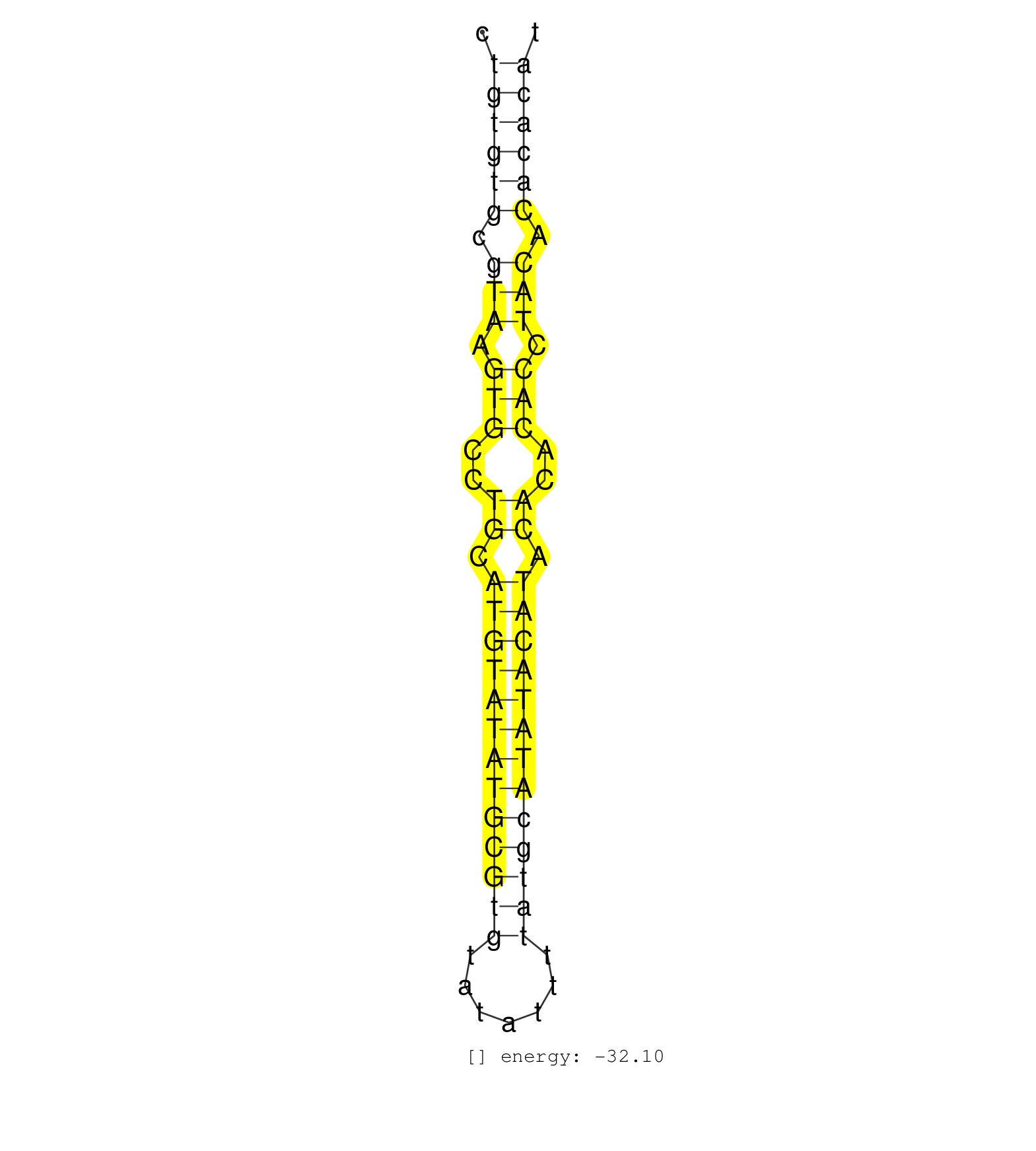
| GAGCAGTTAGAGATCATCCTGGAATAATGTGATGGCCCCTATGTACGTGCCTGTGTGCGTAAGTGCCTGCATGTATATGCGTGTATATTTTATGCATATACATACACACACCTACACACACATGCACACAGACATGCGGGAATAGCCCTCACGGTCCATTTGAATGATATGTG ...................................................((((((.(((.(((..((.(((((((((((((.......))))))))))))).))..))).))).))))))................................................... ..................................................51......................................................................123................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR059765(GSM562827) DN3_control. (thymus) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR059768(GSM562830) Treg_control. (spleen) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................ATGTACGTGCCTGTGTGCG.................................................................................................................. | 19 | 1 | 842.00 | 842.00 | 161.00 | 171.00 | 112.00 | 130.00 | 107.00 | 57.00 | 14.00 | 15.00 | 11.00 | 8.00 | 5.00 | - | 3.00 | 4.00 | 5.00 | 4.00 | 1.00 | 4.00 | 2.00 | 1.00 | 3.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 1.00 | 1.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TGTACGTGCCTGTGTGCG.................................................................................................................. | 18 | 2 | 122.50 | 122.50 | 42.00 | 13.50 | 21.00 | 10.00 | 18.00 | 6.50 | 3.00 | 0.50 | 2.00 | - | 0.50 | - | 0.50 | 0.50 | - | - | - | - | 0.50 | - | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| .......................................TATGTACGTGCCTGTGTGCG.................................................................................................................. | 20 | 1 | 108.00 | 108.00 | 46.00 | 9.00 | 20.00 | 7.00 | 3.00 | 8.00 | 3.00 | - | - | 2.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................CTATGTACGTGCCTGTGTGC................................................................................................................... | 20 | 1 | 66.00 | 66.00 | 12.00 | 11.00 | 13.00 | 10.00 | 10.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TATGTACGTGCCTGTGTGC................................................................................................................... | 19 | 2 | 38.50 | 38.50 | 9.00 | 4.00 | 6.00 | 7.50 | 7.00 | 2.50 | - | 1.00 | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................CTATGTACGTGCCTGTGTGCG.................................................................................................................. | 21 | 1 | 27.00 | 27.00 | 2.00 | 4.00 | 5.00 | 2.00 | 7.00 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATGTACGTGCCTGTGTGC................................................................................................................... | 18 | 6 | 25.83 | 25.83 | 5.17 | 4.17 | 3.00 | 2.50 | 6.83 | 1.50 | 0.33 | 0.17 | - | - | - | - | - | 0.17 | 0.17 | 0.17 | - | - | 0.50 | 0.17 | - | - | - | - | - | 0.17 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | 0.17 | - | - | - | - | - | - | - |
| .....................................CCTATGTACGTGCCTGTGTGC................................................................................................................... | 21 | 1 | 18.00 | 18.00 | - | 3.00 | 4.00 | 2.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGGA................................ | 25 | 10 | 15.80 | 15.80 | - | 0.20 | 0.10 | 0.40 | - | - | 7.90 | - | - | - | - | 4.60 | 0.10 | 0.20 | - | - | 1.80 | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | 0.10 | 0.10 | - | - |
| ..........................................GTACGTGCCTGTGTGCG.................................................................................................................. | 17 | 2 | 9.50 | 9.50 | 3.50 | 0.50 | 1.00 | - | 3.00 | - | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................CCTATGTACGTGCCTGTGTGCG.................................................................................................................. | 22 | 1 | 9.00 | 9.00 | - | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGGAA............................... | 26 | 10 | 6.60 | 6.60 | - | 0.10 | - | - | - | - | 3.00 | - | - | - | - | 0.90 | 0.10 | - | 0.10 | - | 1.60 | - | - | - | - | - | - | - | 0.20 | - | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | 0.10 |
| ....................................CCCTATGTACGTGCCTGTGTGC................................................................................................................... | 22 | 1 | 4.00 | 4.00 | - | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATGTACGTGCCTGTGggcg.................................................................................................................. | 19 | GGCG | 4.00 | 0.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TGTACGTGCCTGTGTGC................................................................................................................... | 17 | 10 | 2.50 | 2.50 | 0.40 | 0.70 | 0.60 | 0.50 | 0.10 | - | - | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATGTACGTGCCTGTGcgcg.................................................................................................................. | 19 | CGCG | 2.00 | 0.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................CCCTATGTACGTGCCTGTGTGCG.................................................................................................................. | 23 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CCCCTATGTACGTGCCTGTGTGC................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATGTACGTGCCTGTGTtcg.................................................................................................................. | 19 | TCG | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGG................................. | 24 | 10 | 1.20 | 1.20 | - | - | - | - | 0.50 | - | 0.20 | - | - | - | - | 0.40 | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATGTACGTGCCTGTGTG.................................................................................................................... | 17 | 9 | 1.11 | 1.11 | 0.33 | 0.67 | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATGTACGTGCCTGTGggc................................................................................................................... | 18 | GGC | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................CTATGTACGTGCCTGTGcgcg.................................................................................................................. | 21 | CGCG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................ATGTACGTGCCTGTGagcg.................................................................................................................. | 19 | AGCG | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GCGGGAATAGCCCTCggtt................... | 19 | GGTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................CTATGTACGTGCCTGTGTacg.................................................................................................................. | 21 | ACG | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................CCTATGTACGTGCCTGTGTGCa.................................................................................................................. | 22 | A | 1.00 | 18.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................CTATGTACGTGCCTGTGggc................................................................................................................... | 20 | GGC | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................GCCCCTATGTACGTGCCTGTGTGCG.................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TATGTACGTGCCTGTGTGCt.................................................................................................................. | 20 | T | 0.50 | 38.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TATGTACGTGCCTGTGTG.................................................................................................................... | 18 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGGAAa.............................. | 27 | A | 0.30 | 6.60 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | 0.10 | - | - | - | - |
| .....................................................................................................................ACACATGCACACAGACATGCGGGAAa.............................. | 26 | A | 0.20 | 0.00 | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGGt................................ | 25 | T | 0.10 | 1.20 | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGGAAT.............................. | 27 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGGAAaa............................. | 28 | AA | 0.10 | 6.60 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGGAt............................... | 26 | T | 0.10 | 15.80 | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CACACATGCACACAGACATGCGGGAc............................... | 26 | C | 0.10 | 15.80 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - |
| GAGCAGTTAGAGATCATCCTGGAATAATGTGATGGCCCCTATGTACGTGCCTGTGTGCGTAAGTGCCTGCATGTATATGCGTGTATATTTTATGCATATACATACACACACCTACACACACATGCACACAGACATGCGGGAATAGCCCTCACGGTCCATTTGAATGATATGTG ...................................................((((((.(((.(((..((.(((((((((((((.......))))))))))))).))..))).))).))))))................................................... ..................................................51......................................................................123................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR059765(GSM562827) DN3_control. (thymus) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (blood) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR059768(GSM562830) Treg_control. (spleen) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|