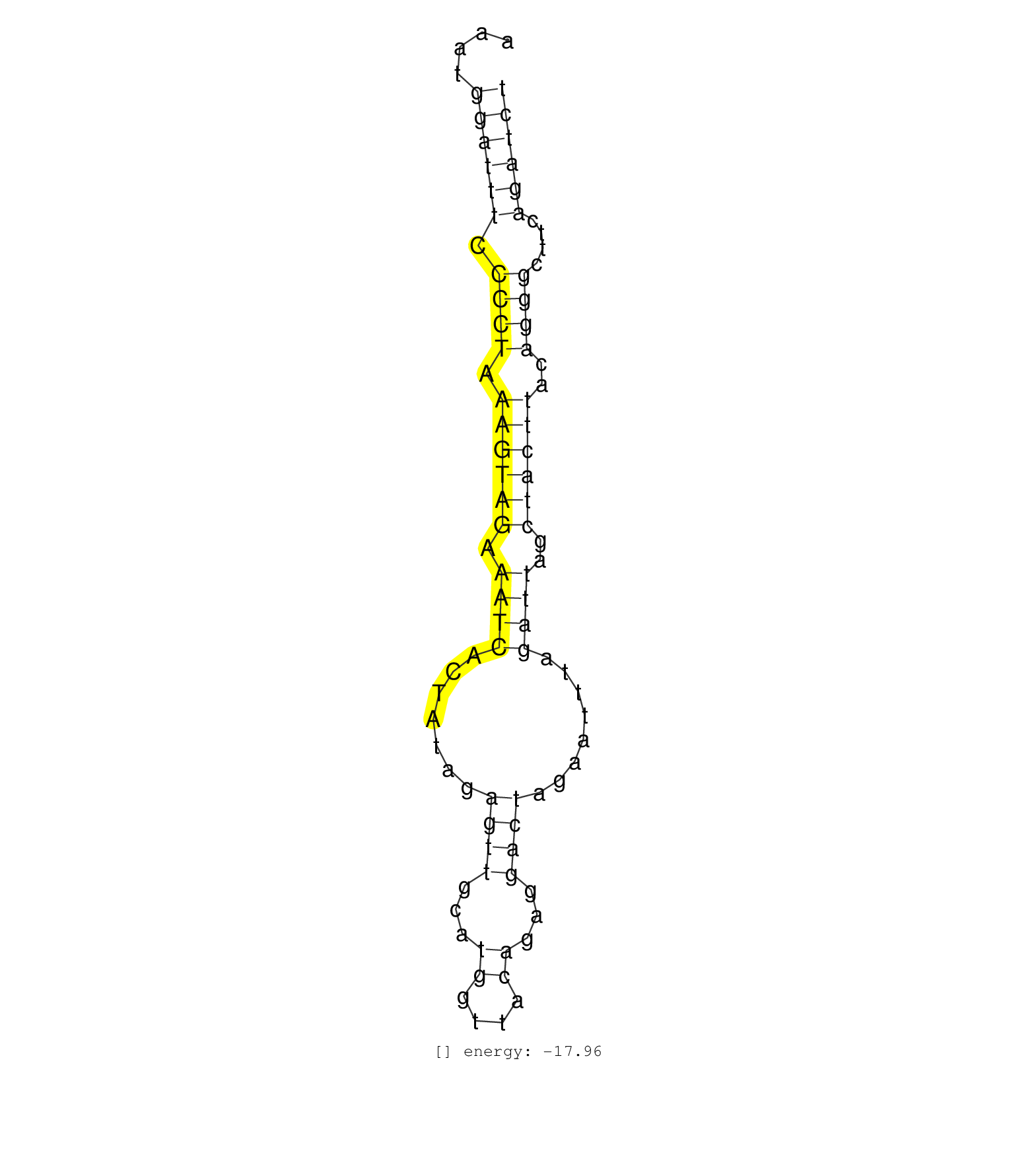
| (1) AGO2.ip | (8) B-CELL | (10) BRAIN | (1) CELL-LINE | (1) DGCR8.mut | (3) EMBRYO | (1) ESC | (3) LIVER | (4) LYMPH | (7) OTHER | (3) OTHER.mut | (1) OVARY | (1) PIWI.mut | (5) SPLEEN | (4) TESTES | (2) THYMUS |
| GACTTTAAAAAGAGATATTCACTTACTAAATGATGGCAAAATATTATAGGAAATGGATTTCCCCTAAAGTAGAAATCACTATAGAGTTGCATGGTTACAGAGGACTAGAATTTAGATTAGCTACTTACAGGGCTTCAGATCTACAGAGTTCTACTTGAGCCCCAGAACCTAGATACTGAAGGCGCTAAGATC ......................................................((((((.((((.((((((.((((.((....)).....(((((......))))).......))))..))))))..))))....)))))).................................................. ..................................................51.........................................................................................142................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR073954(GSM629280) total RNA. (blood) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR073955(GSM629281) total RNA. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR059765(GSM562827) DN3_control. (thymus) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR059768(GSM562830) Treg_control. (spleen) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR065046(SRR065046) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR059775(GSM562837) MEF_Drosha. (MEF) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR077865(GSM637802) 18-30 nt small RNAs. (liver) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................CCCCTAAAGTAGAAAgcac................................................................................................................. | 19 | GCAC | 52.00 | 0.00 | 1.00 | 7.00 | 3.00 | 2.00 | - | 3.00 | - | 3.00 | 4.00 | 2.00 | 5.00 | 3.00 | 1.00 | 2.00 | - | 3.00 | 1.00 | 2.00 | - | 2.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAAgca.................................................................................................................. | 18 | GCA | 27.00 | 0.00 | 11.00 | 2.00 | 1.00 | 2.00 | 2.00 | 2.00 | 1.00 | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAATCACTA............................................................................................................... | 21 | 1 | 11.00 | 11.00 | 2.00 | - | 3.00 | - | - | - | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAATCACTAc.............................................................................................................. | 22 | C | 10.00 | 11.00 | 8.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTAAAGTAGAAATCACTA............................................................................................................... | 19 | 1 | 8.00 | 8.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAAgc................................................................................................................... | 17 | GC | 8.00 | 0.00 | 6.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTAAAGTAGAAATCACTAct............................................................................................................. | 21 | CT | 7.00 | 8.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAAgcaa................................................................................................................. | 19 | GCAA | 6.00 | 0.00 | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAATCACT................................................................................................................ | 20 | 1 | 5.00 | 5.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAAgccc................................................................................................................. | 19 | GCCC | 4.00 | 0.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TAAAGTAGAAATCACTAct............................................................................................................. | 19 | CT | 3.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TAAAGTAGAAATCACTActaa........................................................................................................... | 21 | CTAA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTAAAGTAGAAATCACTAc.............................................................................................................. | 20 | C | 2.00 | 8.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TAAAGTAGAAATCACTActat........................................................................................................... | 21 | CTAT | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTAAAGTAGAAATCACT................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTAAAGTAGAAATCAgta............................................................................................................... | 19 | GTA | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAAgcc.................................................................................................................. | 18 | GCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................CCTAAAGTAGAAATCACTActat........................................................................................................... | 23 | CTAT | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAAgcgc................................................................................................................. | 19 | GCGC | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAATCACTAa.............................................................................................................. | 22 | A | 1.00 | 11.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAATCAat................................................................................................................ | 20 | AT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTAAAGTAGAAATCACaa............................................................................................................... | 19 | AA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAAgac.................................................................................................................. | 18 | GAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GAGTTGCATGGTTACAaatt......................................................................................... | 20 | AATT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CCTAAAGTAGAAATCACTATt............................................................................................................. | 21 | T | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAAGTAGAAATCACTgc.............................................................................................................. | 17 | GC | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................ACAGAGTTCTACTTGgcg................................ | 18 | GCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................CCCTAAAGTAGAAATCACT................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCCCCTAAAGTAGAAAgcc.................................................................................................................. | 19 | GCC | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCCCTAAAGTAGAAATCcct................................................................................................................ | 20 | CCT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................AAAGTAGAAATCACTAct............................................................................................................. | 18 | CT | 0.20 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | 0.10 |
| .................................................................AAAGTAGAAATCACTA............................................................................................................... | 16 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - |
| GACTTTAAAAAGAGATATTCACTTACTAAATGATGGCAAAATATTATAGGAAATGGATTTCCCCTAAAGTAGAAATCACTATAGAGTTGCATGGTTACAGAGGACTAGAATTTAGATTAGCTACTTACAGGGCTTCAGATCTACAGAGTTCTACTTGAGCCCCAGAACCTAGATACTGAAGGCGCTAAGATC ......................................................((((((.((((.((((((.((((.((....)).....(((((......))))).......))))..))))))..))))....)))))).................................................. ..................................................51.........................................................................................142................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR073954(GSM629280) total RNA. (blood) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR073955(GSM629281) total RNA. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR059765(GSM562827) DN3_control. (thymus) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR059768(GSM562830) Treg_control. (spleen) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR065046(SRR065046) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR059775(GSM562837) MEF_Drosha. (MEF) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR077865(GSM637802) 18-30 nt small RNAs. (liver) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................cccaACTAAATGATGGCAA......................................................................................................................................................... | 19 | ccca | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......aagAGAGATATTCACTTAC...................................................................................................................................................................... | 19 | aag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................cagaTTCAGATCTACAGAGT........................................... | 20 | caga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................aaccACAGGGCTTCAGATCT.................................................. | 20 | aacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................tccaTAAAGTAGAAATCACTA............................................................................................................... | 21 | tcca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................taaaGTTACAGAGGACTAGA................................................................................... | 20 | taaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |