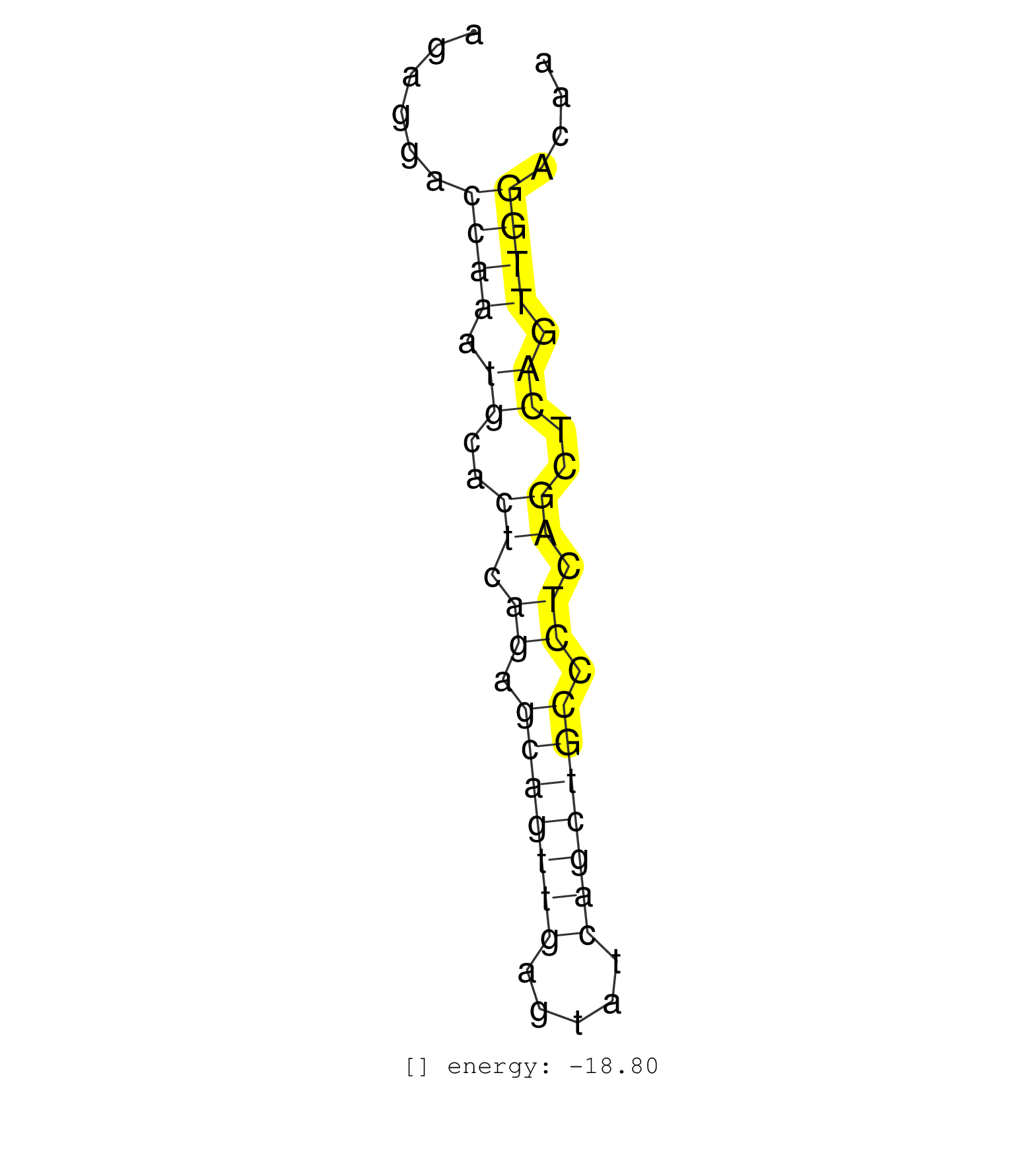
| (7) AGO2.ip | (10) BRAIN | (1) CELL-LINE | (2) ESC | (1) LIVER | (2) SPLEEN |
| GACTCCTAGTTGGCCTGCTCCCATGAAGATTCCATATCCCGATATATCCTAGAGGACCAAATGCACTCAGAGCAGTTGAGTATCAGCTGCCCTCAGCTCAGTTGGACAACAACTTGACTACTCCACCTAAAGACTCAACAGTCTGAAGGCCAACCAGGA ........................................................((((.((..((.((.(((((((.....))))))).)).))..)).))))...................................................... ..................................................51........................................................109................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR037938(GSM510476) 293Red. (cell line) | SRR059771(GSM562833) CD4_control. (spleen) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................CCCTCAGCTCAGTTGGA..................................................... | 17 | 1 | 6.00 | 6.00 | 2.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................CCTCAGCTCAGTTGGAtaga................................................. | 20 | TAGA | 5.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................CCTCAGCTCAGTTGGA..................................................... | 16 | 1 | 4.00 | 4.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................GCCCTCAGCTCAGTTGGA..................................................... | 18 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................CCTCAGCTCAGTTGGAt.................................................... | 17 | T | 3.00 | 4.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CCCTCAGCTCAGTTGGAt.................................................... | 18 | T | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GCCCTCAGCTCAGTTGGAtagc................................................. | 22 | TAGC | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CCTCAGCTCAGTTGGAta................................................... | 18 | TA | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGCCCTCAGCTCAGTTGGAt.................................................... | 20 | T | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................CCCTCAGCTCAGTTGGAC.................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................CAGCTGCCCTCAGCTCActgt....................................................... | 21 | CTGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................CCCTCAGCTCAGTTGGc..................................................... | 17 | C | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................CAGAGCAGTTGAGTAga........................................................................... | 17 | GA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................TCAGCTCAGTTGGACA................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - |
| GACTCCTAGTTGGCCTGCTCCCATGAAGATTCCATATCCCGATATATCCTAGAGGACCAAATGCACTCAGAGCAGTTGAGTATCAGCTGCCCTCAGCTCAGTTGGACAACAACTTGACTACTCCACCTAAAGACTCAACAGTCTGAAGGCCAACCAGGA ........................................................((((.((..((.((.(((((((.....))))))).)).))..)).))))...................................................... ..................................................51........................................................109................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR037938(GSM510476) 293Red. (cell line) | SRR059771(GSM562833) CD4_control. (spleen) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................gcGCCCTCAGCTCAGTTG....................................................... | 18 | gc | 4.00 | 0.00 | - | - | - | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ........gtcaGCCTGCTCCCATGAA.................................................................................................................................... | 19 | gtca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........aaaaGCCTGCTCCCATGAA.................................................................................................................................... | 19 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............atagCCCATGAAGATTCCA............................................................................................................................. | 19 | atag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |