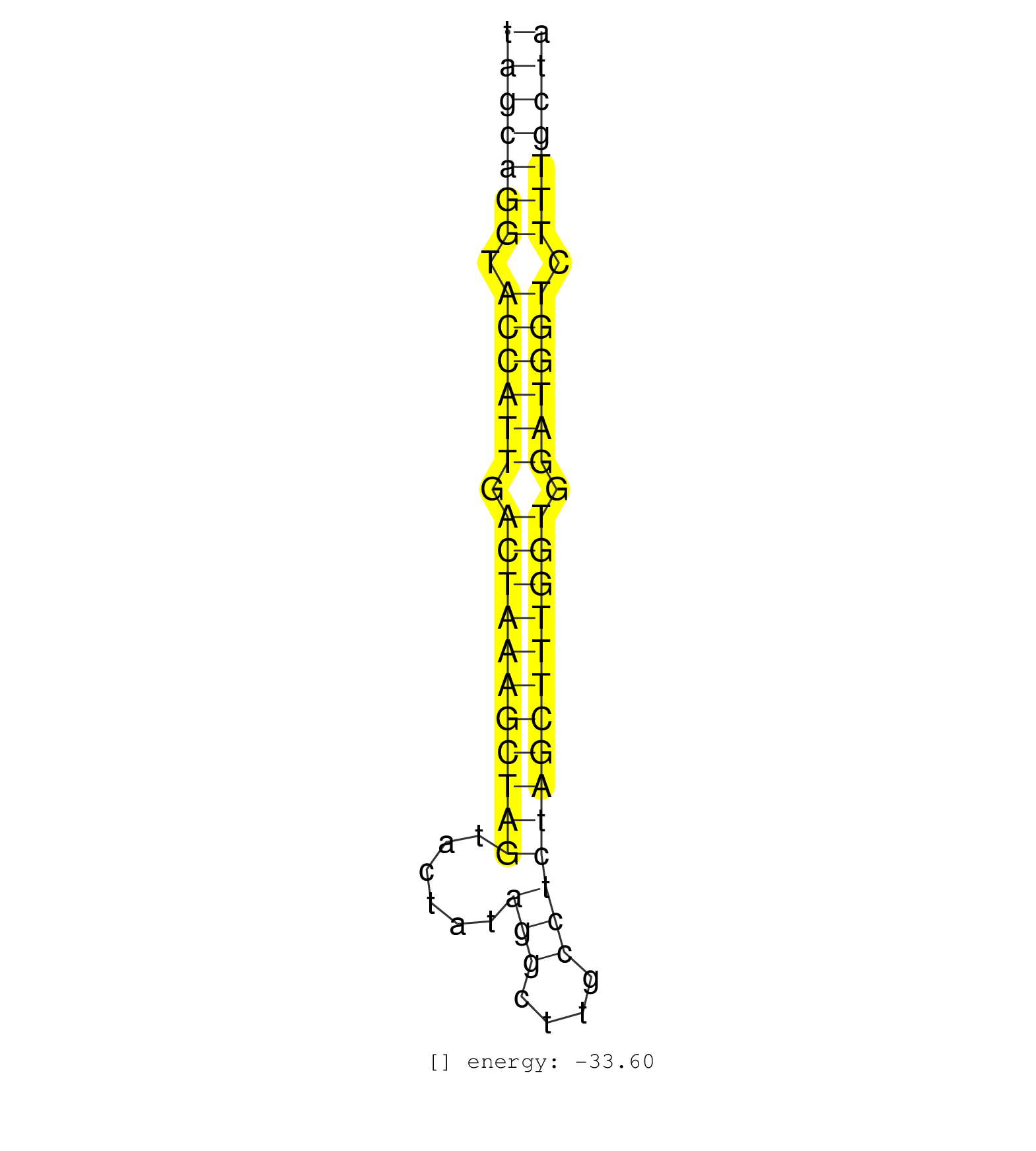
| (2) AGO1.ip | (20) AGO2.ip | (8) B-CELL | (40) BRAIN | (2) CELL-LINE | (2) DGCR8.mut | (15) EMBRYO | (6) ESC | (6) FIBROBLAST | (5) LIVER | (4) LYMPH | (10) OTHER | (11) OTHER.mut | (1) OVARY | (1) PIWI.ip | (3) SKIN | (8) SPLEEN | (18) TESTES | (3) THYMUS | (10) TOTAL-RNA | (1) UTERUS |
| GGACCCTGGAACTGCGGATGTGGTCATGGTGTGGGTCAGTGGGGCAGCAGGTGGGGTAGGTAGCAGGTACCATTGACTAAAGCTAGTACTATAGGCTTGCCTCTAGCTTTGGTGGATGGTCTTTGCTAAGTTCTCTGCCAACCCAGCAGTGAGAATTGGGGTACAGAAAAGGAGGAAGAGGTCTGCAG ............................................................(((((((.((((((.(((((((((((......(((....)))))))))))))).)))))).)))))))............................................................ ............................................................61.................................................................128.......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR073954(GSM629280) total RNA. (blood) | GSM510444(GSM510444) brain_rep5. (brain) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR037928(GSM510466) e7p5_rep2. (embryo) | mjTestesWT4() Testes Data. (testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR037921(GSM510459) e12p5_rep3. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR206940(GSM723281) other. (brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR306528(GSM750571) 19-24nt. (ago2 brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR037905(GSM510441) brain_rep2. (brain) | SRR037909(GSM510446) newborn_rep2. (total RNA) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR059767(GSM562829) DN3_Dicer. (thymus) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR014234(GSM319958) Ovary total. (ovary) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | GSM475281(GSM475281) total RNA. (testes) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR206941(GSM723282) other. (brain) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | GSM640583(GSM640583) small RNA in the liver with paternal Low prot. (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR206939(GSM723280) other. (brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (testes) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR060845(GSM561991) total RNA. (brain) | SRR059765(GSM562827) DN3_control. (thymus) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM510453(GSM510453) newborn_rep9. (total RNA) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | SRR073955(GSM629281) total RNA. (blood) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | GSM510452(GSM510452) newborn_rep8. (total RNA) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR037910(GSM510447) newborn_rep3. (total RNA) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037908(GSM510445) newborn_rep1. (total RNA) | GSM261957(GSM261957) oocytesmallRNA-19to24. (oocyte) | SRR206942(GSM723283) other. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................GGTACCATTGACTAAAGCTAG...................................................................................................... | 21 | 1 | 457.00 | 457.00 | 44.00 | 29.00 | 23.00 | 24.00 | 19.00 | 24.00 | 18.00 | 16.00 | 17.00 | 14.00 | 16.00 | 10.00 | 2.00 | 17.00 | 9.00 | 16.00 | 4.00 | - | 6.00 | 7.00 | 10.00 | - | 7.00 | 7.00 | 4.00 | 6.00 | - | 8.00 | 7.00 | 6.00 | 6.00 | 4.00 | 5.00 | 2.00 | 4.00 | - | 3.00 | - | 1.00 | 4.00 | - | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | 3.00 | - | 3.00 | 3.00 | 2.00 | - | 4.00 | - | - | 2.00 | - | 3.00 | - | - | - | - | - | - | 1.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | 2.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGt.............................................................. | 22 | T | 35.00 | 6.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 3.00 | - | 3.00 | - | - | 4.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCT........................................................................................................ | 19 | 1 | 29.00 | 29.00 | 1.00 | - | 1.00 | - | 2.00 | - | 1.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTT................................................................ | 20 | 1 | 22.00 | 22.00 | - | - | - | - | - | - | - | - | - | - | - | - | 9.00 | - | - | - | - | 3.00 | - | - | - | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTAGT..................................................................................................... | 22 | 1 | 21.00 | 21.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 2.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTCTTT................................................................ | 21 | 1 | 20.00 | 20.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 3.00 | - | - | - | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTA....................................................................................................... | 20 | 1 | 16.00 | 16.00 | - | 1.00 | 1.00 | 1.00 | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................CTAGCTTTGGTGGATGGTCTTT................................................................ | 22 | 1 | 13.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTAGa..................................................................................................... | 22 | A | 12.00 | 457.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................CAGGTGGGGTAGGTAGCA........................................................................................................................... | 18 | 1 | 12.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AGGTACCATTGACTAAAGCTAG...................................................................................................... | 22 | 1 | 9.00 | 9.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................ACCATTGACTAAAGCTAG...................................................................................................... | 18 | 1 | 9.00 | 9.00 | - | - | - | - | 1.00 | - | - | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGa.............................................................. | 22 | A | 8.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTCTTTG............................................................... | 22 | 1 | 7.00 | 7.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTG............................................................... | 21 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTCTTTGt.............................................................. | 23 | T | 5.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CAGGTACCATTGACTAAAGCT........................................................................................................ | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AGGTACCATTGACTAAAGCTA....................................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................GCAGGTGGGGTAGGTAGCA........................................................................................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTAGTA.................................................................................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCT.................................................................. | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTAGc..................................................................................................... | 22 | C | 3.00 | 457.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................AAAAGGAGGAAGAGGTCTGCAGgata | 26 | GATA | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTT................................................................. | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTAt...................................................................................................... | 21 | T | 3.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTCTT................................................................. | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................GTACCATTGACTAAAGCTAG...................................................................................................... | 20 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CTAAGTTCTCTGCCAACC............................................. | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGC......................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGtaa............................................................ | 24 | TAA | 2.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CTCTAGCTTTGGTGGATGGTCTag................................................................ | 24 | AG | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................AGCAGGTGGGGTAGGTAGCA........................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CAGGTACCATTGACTAAAGCTAGa..................................................................................................... | 24 | A | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTCTTTGa.............................................................. | 23 | A | 2.00 | 7.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGGTCATGGTGTGGGTCAGTGGGGCAGCAGGTG....................................................................................................................................... | 33 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTC................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTCTTTaa.............................................................. | 23 | AA | 2.00 | 20.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AGGTACCATTGACTAAAGCTAGa..................................................................................................... | 23 | A | 2.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................TGGGGCAGCAGGTGGtggg.................................................................................................................................. | 19 | TGGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTtt.............................................................. | 22 | TT | 1.00 | 22.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TTTGGTGGATGGTCTTg................................................................ | 17 | G | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TACCATTGACTAAAGCTA....................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................GGAGGAAGAGGTCTGCtac | 19 | TAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTattt................................................................ | 20 | ATTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................CTTTGGTGGATGGTCTTTG............................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGCT............................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTgg...................................................................................................... | 21 | GG | 1.00 | 29.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TTTGGTGGATGGTCTTTGt.............................................................. | 19 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................GGAGGAAGAGGTCTGtcct | 19 | TCCT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CCATTGACTAAAGCTAG...................................................................................................... | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CAGGTGGGGTAGGTAGCAGG......................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTcg...................................................................................................... | 21 | CG | 1.00 | 29.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTAGCTTTGGTGGATGGTCTTTt............................................................... | 23 | T | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGGGGCAGCAGGTGGaca................................................................................................................................... | 18 | ACA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTAGCTTTGGTGGATGGTCTTTat.............................................................. | 24 | AT | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGGGGCAGCAGGTGGcatc.................................................................................................................................. | 19 | CATC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TTGGTGGATGGTCTTTGCTAAG.......................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGGGGCAGCAGGTGGtgtg.................................................................................................................................. | 19 | TGTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGGAACTGCGGATGTGGTCATGGT.............................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTtaa............................................................. | 23 | TAA | 1.00 | 22.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................ACAGAAAAGGAGGAAGttg....... | 19 | TTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GCAGCAGGTGGGGTAGGTAGCA........................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGaa............................................................. | 23 | AA | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTat.............................................................. | 22 | AT | 1.00 | 22.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTt............................................................... | 21 | T | 1.00 | 22.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTCTTTGttt............................................................ | 25 | TTT | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTAGCTTTGGTGGATGGTCT.................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................GTTCTCTGCCAACCCccaa........................................ | 19 | CCAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTAGCTTTGGTGGATGGTCTTa................................................................ | 22 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGGGGCAGCAGGTGGttta.................................................................................................................................. | 19 | TTTA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGga............................................................. | 23 | GA | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGCTAa...................................................................................................... | 21 | A | 1.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGttt............................................................ | 24 | TTT | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGTACCATTGACTAAAGgt........................................................................................................ | 19 | GT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................AGGTACCATTGACTAAAGCT........................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................CAGCAGGTGGGGTAGGTAGCA........................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGGGGCAGCAGGTGGtaca.................................................................................................................................. | 19 | TACA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CAGGTACCATTGACTAAAGCTA....................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTGGGGTAGGTAGCAGGT........................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TTTGGTGGATGGTCTTTGCTA............................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGtttt........................................................... | 25 | TTTT | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GCAGGTACCATTGACcgga........................................................................................................... | 19 | CGGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGGAACTGCGGATGTGGTCATGGTGT............................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CTGGAACTGCGGATGTGGTCATGGTG............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGGGGCAGCAGGTGGtgt................................................................................................................................... | 18 | TGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................AGCTTTGGTGGATGGTCTTTGC.............................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGCTTTGGTGGATGGTCTTTGat............................................................. | 24 | AT | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GCTTTGGTGGATGGTCTT................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GCTTTGGTGGATGGTCTTT................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGGGTCAGTGGGGCAGggag......................................................................................................................................... | 20 | GGAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CCATTGACTAAAGCTAGT..................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGACCCTGGAACTGCGGATGTGGTCATGGTGTGGGTCAGTGGGGCAGCAGGTGGGGTAGGTAGCAGGTACCATTGACTAAAGCTAGTACTATAGGCTTGCCTCTAGCTTTGGTGGATGGTCTTTGCTAAGTTCTCTGCCAACCCAGCAGTGAGAATTGGGGTACAGAAAAGGAGGAAGAGGTCTGCAG ............................................................(((((((.((((((.(((((((((((......(((....)))))))))))))).)))))).)))))))............................................................ ............................................................61.................................................................128.......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR073954(GSM629280) total RNA. (blood) | GSM510444(GSM510444) brain_rep5. (brain) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR037928(GSM510466) e7p5_rep2. (embryo) | mjTestesWT4() Testes Data. (testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR037921(GSM510459) e12p5_rep3. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR206940(GSM723281) other. (brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR306528(GSM750571) 19-24nt. (ago2 brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR037905(GSM510441) brain_rep2. (brain) | SRR037909(GSM510446) newborn_rep2. (total RNA) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR059767(GSM562829) DN3_Dicer. (thymus) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR014234(GSM319958) Ovary total. (ovary) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | GSM475281(GSM475281) total RNA. (testes) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR206941(GSM723282) other. (brain) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | GSM640583(GSM640583) small RNA in the liver with paternal Low prot. (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR206939(GSM723280) other. (brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (testes) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR060845(GSM561991) total RNA. (brain) | SRR059765(GSM562827) DN3_control. (thymus) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM510453(GSM510453) newborn_rep9. (total RNA) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | SRR073955(GSM629281) total RNA. (blood) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | GSM510452(GSM510452) newborn_rep8. (total RNA) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR037910(GSM510447) newborn_rep3. (total RNA) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037908(GSM510445) newborn_rep1. (total RNA) | GSM261957(GSM261957) oocytesmallRNA-19to24. (oocyte) | SRR206942(GSM723283) other. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) |
|---|