| (1) AGO1.ip | (17) AGO2.ip | (2) AGO3.ip | (6) B-CELL | (31) BRAIN | (10) CELL-LINE | (1) DGCR8.mut | (14) EMBRYO | (6) ESC | (8) FIBROBLAST | (7) LIVER | (4) LYMPH | (5) OTHER | (3) OTHER.mut | (1) OVARY | (1) PANCREAS | (2) PIWI.ip | (3) SKIN | (1) SPLEEN | (7) TESTES | (1) THYMUS | (3) TOTAL-RNA |
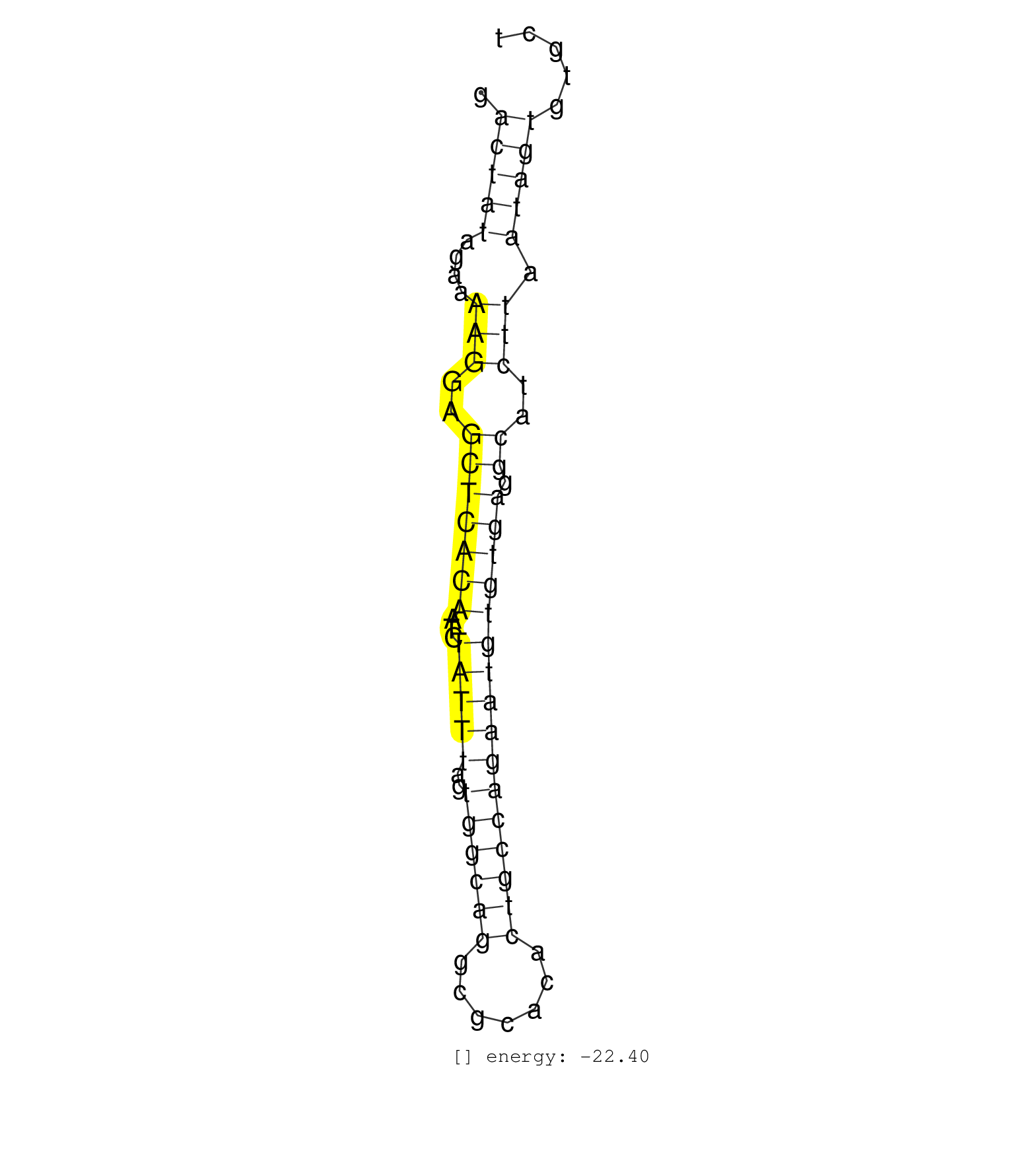
| TTAAGTTAAGATCAGCCAAACAAGATCTTGCTGGCTCTTCTGGTCACATGGACTATAGAAAAGGAGCTCACAATCTATTTAGTGGCAGGCGCACACTGCCAGAATGTGTGAGGCATCTTAATAGTGTGCTGATGACACTGTTTTGGAAAGGAAGGTTTGTAACACTTGGAGGTGGGGGGT ...................................................(((((....(((..(((((((...(((((..((((((.......)))))))))))))))).))..))).)))))....................................................... ..................................................51.............................................................................130................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR390298(GSM849856) cell line: NIH-3T3cell type: fibroblastinfect. (fibroblast) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR059765(GSM562827) DN3_control. (thymus) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR037938(GSM510476) 293Red. (cell line) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR206939(GSM723280) other. (brain) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR037935(GSM510473) 293cand3. (cell line) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR037922(GSM510460) e12p5_rep4. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR206942(GSM723283) other. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR065049(SRR065049) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | mjTestesWT3() Testes Data. (testes) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR037936(GSM510474) 293cand1. (cell line) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR065047(SRR065047) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR014234(GSM319958) Ovary total. (ovary) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR037907(GSM510443) brain_rep4. (brain) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037926(GSM510464) e9p5_rep4. (embryo) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR037905(GSM510441) brain_rep2. (brain) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR306528(GSM750571) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................AAGGAGCTCACAATCTATTgag.................................................................................................. | 22 | GAG | 231.00 | 4.00 | 29.00 | 33.00 | 29.00 | 17.00 | 16.00 | 7.00 | - | 8.00 | 7.00 | - | 7.00 | 11.00 | - | - | - | - | 7.00 | 6.00 | 7.00 | 1.00 | 5.00 | 6.00 | - | 1.00 | - | - | 4.00 | 1.00 | 2.00 | - | - | 1.00 | - | 1.00 | 3.00 | - | - | - | 3.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgctg................................................................................................... | 21 | GCTG | 47.00 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 9.00 | - | - | - | - | 6.00 | - | - | - | 4.50 | - | - | - | - | 2.50 | 0.50 | 3.00 | 2.00 | - | - | - | - | 2.50 | 3.00 | 0.50 | - | - | - | - | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | 1.00 | 1.00 | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | 0.50 | 0.50 | 0.50 | - |
| ............................................................AAGGAGCTCACAATCTATTga................................................................................................... | 21 | GA | 35.00 | 4.00 | 2.00 | 1.00 | 2.00 | 3.00 | 2.00 | 6.00 | - | - | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgct.................................................................................................... | 20 | GCT | 28.00 | 1.50 | - | - | - | 0.50 | - | - | - | 2.00 | - | 7.50 | 0.50 | - | - | 3.00 | 0.50 | - | - | - | - | - | - | - | - | - | 1.50 | - | - | - | - | 1.50 | 1.00 | - | - | - | - | 1.00 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | 2.50 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTgagt................................................................................................. | 23 | GAGT | 27.00 | 4.00 | 4.00 | - | 1.00 | 2.00 | 6.00 | - | - | 5.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTgaga................................................................................................. | 23 | GAGA | 15.00 | 4.00 | - | 1.00 | - | 1.00 | 2.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgc..................................................................................................... | 19 | GC | 7.50 | 1.50 | - | - | - | - | - | - | - | - | 1.50 | 1.00 | 0.50 | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTg.................................................................................................... | 20 | G | 7.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTgaa.................................................................................................. | 22 | GAA | 4.00 | 4.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATT..................................................................................................... | 19 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTcag.................................................................................................. | 22 | CAG | 3.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgcc.................................................................................................... | 20 | GCC | 3.00 | 1.50 | - | - | - | - | - | - | - | - | - | 1.50 | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAc...................................................................................................... | 18 | C | 2.50 | 1.50 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgcta................................................................................................... | 21 | GCTA | 2.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTgat.................................................................................................. | 22 | GAT | 2.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GGAGCTCACAATCTAgct.................................................................................................... | 18 | GCT | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGGAGCTCACAATCTAgta.................................................................................................... | 19 | GTA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgca.................................................................................................... | 20 | GCA | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ............................................................AAGGAGCTCACAATCTA....................................................................................................... | 17 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGGAGCTCACAATCTAgctg................................................................................................... | 20 | GCTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATac.................................................................................................... | 20 | AC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAAGGAGCTCACAATggac...................................................................................................... | 20 | GGAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GGAGCTCACAATCTAgtat................................................................................................... | 19 | GTAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTgca.................................................................................................. | 22 | GCA | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATc..................................................................................................... | 19 | C | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgcg.................................................................................................... | 20 | GCG | 1.00 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGGAGCTCACAATCTAgtg.................................................................................................... | 19 | GTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATagag.................................................................................................. | 22 | AGAG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GGAGCTCACAATCTATTTtg.................................................................................................. | 20 | TG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGGAGCTCACAATCTATTga................................................................................................... | 20 | GA | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATctgg.................................................................................................. | 22 | CTGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................ATGGACTATAGAAAAGGAttt................................................................................................................ | 21 | TTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATctga.................................................................................................. | 22 | CTGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GGAGCTCACAATCTAgtg.................................................................................................... | 18 | GTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAcca.................................................................................................... | 20 | CCA | 1.00 | 1.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................ACTGTTTTGGAAAGGAAagt........................ | 20 | AGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGGAGCTCACAATCTATTgag.................................................................................................. | 21 | GAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTTcgg................................................................................................. | 23 | CGG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTgaaa................................................................................................. | 23 | GAAA | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATcgcg.................................................................................................. | 22 | CGCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCgag...................................................................................................... | 18 | GAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTATTTAt.................................................................................................. | 22 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAct..................................................................................................... | 19 | CT | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAcct.................................................................................................... | 20 | CCT | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgaa.................................................................................................... | 20 | GAA | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgcct................................................................................................... | 21 | GCCT | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgtaa................................................................................................... | 21 | GTAA | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAgta.................................................................................................... | 20 | GTA | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTAcc..................................................................................................... | 19 | CC | 0.50 | 1.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTggct.................................................................................................... | 20 | GGCT | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCT........................................................................................................ | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTcg...................................................................................................... | 18 | CG | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................AAGGAGCTCACAATCTtg...................................................................................................... | 18 | TG | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTAAGTTAAGATCAGCCAAACAAGATCTTGCTGGCTCTTCTGGTCACATGGACTATAGAAAAGGAGCTCACAATCTATTTAGTGGCAGGCGCACACTGCCAGAATGTGTGAGGCATCTTAATAGTGTGCTGATGACACTGTTTTGGAAAGGAAGGTTTGTAACACTTGGAGGTGGGGGGT ...................................................(((((....(((..(((((((...(((((..((((((.......)))))))))))))))).))..))).)))))....................................................... ..................................................51.............................................................................130................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR390298(GSM849856) cell line: NIH-3T3cell type: fibroblastinfect. (fibroblast) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (liver) | SRR059765(GSM562827) DN3_control. (thymus) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR037938(GSM510476) 293Red. (cell line) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR206939(GSM723280) other. (brain) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR037935(GSM510473) 293cand3. (cell line) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR037922(GSM510460) e12p5_rep4. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR206942(GSM723283) other. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR065049(SRR065049) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | mjTestesWT3() Testes Data. (testes) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR037936(GSM510474) 293cand1. (cell line) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR065047(SRR065047) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR014234(GSM319958) Ovary total. (ovary) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR037907(GSM510443) brain_rep4. (brain) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037926(GSM510464) e9p5_rep4. (embryo) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR037905(GSM510441) brain_rep2. (brain) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR306528(GSM750571) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................tcctGCTCACAATCTATTT.................................................................................................... | 19 | tcct | 44.00 | 0.00 | - | - | - | - | - | - | 12.00 | - | - | - | - | - | 7.00 | - | - | 7.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................cctGCTCACAATCTATTT.................................................................................................... | 18 | cct | 21.00 | 0.00 | - | - | - | - | - | - | 7.00 | - | - | - | - | - | 4.00 | - | - | 1.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................atcaACTGCCAGAATGTGT....................................................................... | 19 | atca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ATAGAAAAGGAGCTCACAATCTATT..................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................AATAGTGTGCTGATGACACTGTTTT.................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GCAGGCGCACACTGCC................................................................................ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................ggaATGGACTATAGAAAA...................................................................................................................... | 18 | gga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................tccAGGAGCTCACAATCTA....................................................................................................... | 19 | tcc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AAAAGGAGCTCACAATCTATTT.................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................AGAAAAGGAGCTCACAATCT........................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................tcatGCTCACAATCTATTT.................................................................................................... | 19 | tcat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................agcAGCTCACAATCTATT..................................................................................................... | 18 | agc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |