| (5) BRAIN | (2) CELL-LINE | (1) DGCR8.mut | (1) EMBRYO | (2) ESC | (1) KIDNEY | (9) LIVER | (1) OTHER | (4) OTHER.mut | (1) OVARY | (2) PANCREAS | (1) PIWI.ip | (1) PIWI.mut | (1) SPLEEN | (10) TESTES | (1) THYMUS |
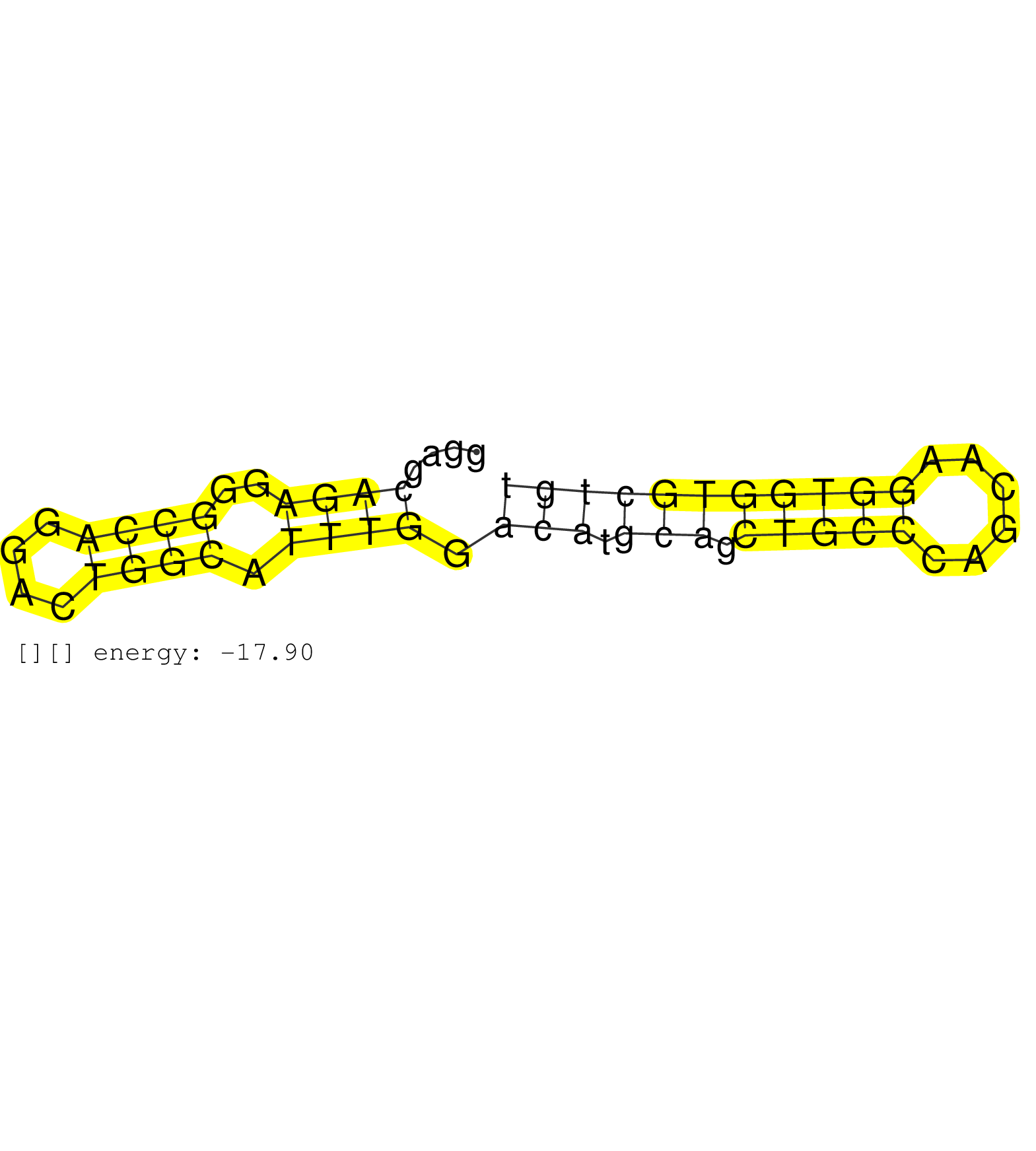
| AATCGGATTCTTGGCATTCCTCTGTCCCTGGCCAGCATGCTCCCCACTGGAGAAGGAGCAGAGGGCCAGGACTGGCATTTGGACATGCAGCTGCCCAGCAAGGTGGTGCTGTCCGCAGCTGCGCTGCTCCTGGTAACTGCGGCTTACAAACTCTACAAGTCCAGACCTGCCCCAGTCGGGCAGGCAGGGAGAAAC ......................................................((((..((((....)))).)))).(((.(((.(((((......))))))))))).................................................. ......................................................55.......................................................112................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | mjLiverKO2() Liver Data. (Zcchc11 liver) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042481(GSM539873) mouse pancreatic tissue [09-002]. (pancreas) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR059765(GSM562827) DN3_control. (thymus) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR065045(SRR065045) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR206942(GSM723283) other. (brain) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | mjLiverWT1() Liver Data. (liver) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR206941(GSM723282) other. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................GAGGGCCAGGACTGGCATTTGGACATGCA.......................................................................................................... | 29 | 1 | 11.00 | 11.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GGGCCAGGACTGGCATTTGGACATGCAG......................................................................................................... | 28 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGGGCCAGGACTGGCATTTGGACATGCAGC........................................................................................................ | 30 | 1 | 4.00 | 4.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GCTGCCCAGCAAGGTGGTG....................................................................................... | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................CCCACTGGAGAAGGAGCA....................................................................................................................................... | 18 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................ATTTGGACATGCAGCTG...................................................................................................... | 17 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CTGCCCAGCAAGGTGGTG....................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................ATTTGGACATGCAGCT....................................................................................................... | 16 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGGGCCAGGACTGGCATT.................................................................................................................... | 18 | 2 | 1.50 | 1.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ...........................................................AGAGGGCCAGGACTGGCATTTGG................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................TTTGGACATGCAGCTGC..................................................................................................... | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CAAGGTGGTGCTGTCCG................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................AGCAAGGTGGTGCTGTCCGCAGat........................................................................... | 24 | AT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CGGGCAGGCAGGGAGtaa. | 18 | TAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................AGGGCCAGGACTGGCATTTGGACATGC........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGAGGGCCAGGACTGGCATTTG.................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GCCAGGACTGGCATTTGGACATGCAGCT....................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................TTGGACATGCAGCTGCCCAGCAAGGTGG......................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GGCCAGGACTGGCATaact................................................................................................................. | 19 | AACT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CTGGAGAAGGAGCAGgtga.................................................................................................................................. | 19 | GTGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................GCAGCTGCCCAGCAAGGT........................................................................................... | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - |
| ......................................GCTCCCCACTGGAGAAcca.......................................................................................................................................... | 19 | CCA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TCTACAAGTCCAGACgatc......................... | 19 | GATC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................GCAAGGTGGTGCTGTCCG................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGAGGGCCAGGACTGGCATTTGGACATGC........................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................ATTTGGACATGCAGCTGCCCAGCAAGGcg.......................................................................................... | 29 | CG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................AGAGGGCCAGGACTGGCATTTGGAg............................................................................................................... | 25 | G | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GCAGAGGGCCAGGACTGGCATT.................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GGCCAGGACTGGCATTaca................................................................................................................. | 19 | ACA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGGAGCAGAGGGCCAccag........................................................................................................................... | 19 | CCAG | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................ACTGGCATTTGGACATGCAGC........................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................AGCAGAGGGCCAGGAggga........................................................................................................................ | 19 | GGGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................ATTTGGACATGCAGCTGCCCA.................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................GGGCCAGGACTGGCATTTGGA................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................ACTGGCATTTGGACATGC........................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................ATTTGGACATGCAGC........................................................................................................ | 15 | 4 | 0.75 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.75 | - | - | - |
| .................................................................CCAGGACTGGCATTTGGA................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TTTGGACATGCAGCTGCCC................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .........................................................GCAGAGGGCCAGGACTGG........................................................................................................................ | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| AATCGGATTCTTGGCATTCCTCTGTCCCTGGCCAGCATGCTCCCCACTGGAGAAGGAGCAGAGGGCCAGGACTGGCATTTGGACATGCAGCTGCCCAGCAAGGTGGTGCTGTCCGCAGCTGCGCTGCTCCTGGTAACTGCGGCTTACAAACTCTACAAGTCCAGACCTGCCCCAGTCGGGCAGGCAGGGAGAAAC ......................................................((((..((((....)))).)))).(((.(((.(((((......))))))))))).................................................. ......................................................55.......................................................112................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | mjLiverKO2() Liver Data. (Zcchc11 liver) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042481(GSM539873) mouse pancreatic tissue [09-002]. (pancreas) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR059765(GSM562827) DN3_control. (thymus) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR065045(SRR065045) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR206942(GSM723283) other. (brain) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | mjLiverWT1() Liver Data. (liver) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR206941(GSM723282) other. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................GCTGCTCCTGGTAACTGCGGCTTACA............................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGCGGCTTACAAACTCTACAAGTCCA................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................GCTGTCCGCAGCTGCGCTGCTCCTGGTA............................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GGTAACTGCGGCTTACAAACTCTACAA..................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................AGCAAGGTGGTGCTGTCCGCAG............................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................ACTCTACAAGTCCAGACCTGCCCCA..................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TTCCTCTGTCCCTGGCCAGCATGCTCC........................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CTGCGGCTTACAAACTCTACAAGTCCA................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................tgtGGCCAGGACTGGCATT.................................................................................................................... | 19 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................ccAACTCTACAAGTCCA................................ | 17 | cc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................ACTGCGGCTTACAAACTCTACAAGTCCA................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |