| (2) AGO2.ip | (2) B-CELL | (16) BRAIN | (1) CELL-LINE | (1) DGCR8.mut | (1) EMBRYO | (1) HEART | (1) KIDNEY | (5) LIVER | (3) LYMPH | (9) OTHER | (2) OTHER.mut | (1) OVARY | (2) SPLEEN | (3) TESTES | (2) THYMUS |
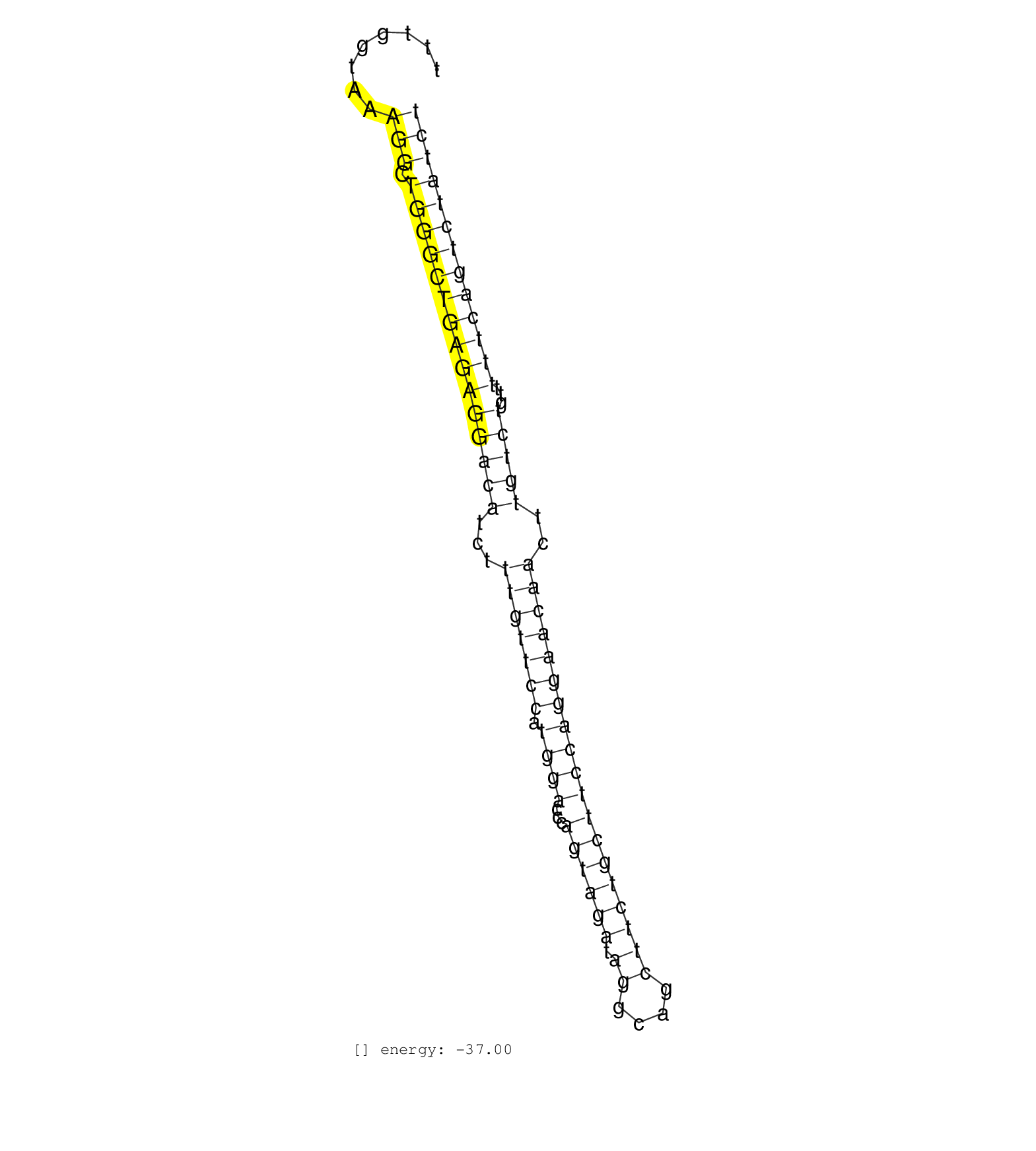
| GTTGGCATCATGTCACTTCAGGTTAGATGTACATGCTCTTGATGTACAGGGCATCATTTGGTAAAGGCTGGGCTGAGAGGACATCTTTGTTCCATGGACCCAGTAGATAGGCAGCTTCTGCTTCCAGGAACAACTTGTCTGTTTTTCAGTCTATCTACTTCCTAAAATTTCAATTTGCTCACATAGCACTTAAGAGAAGAGCTCTC ...........................................................(((....)))((((((((((((((...(((((((.((((...((((((.((....)))))))))))))))))))..)))))...)))))))))...................................................... ........................................................57.................................................................................................156................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | mjLiverWT1() Liver Data. (liver) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR095855BC6(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR059765(GSM562827) DN3_control. (thymus) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | mjTestesWT4() Testes Data. (testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR206940(GSM723281) other. (brain) | SRR073954(GSM629280) total RNA. (blood) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR037937(GSM510475) 293cand2. (cell line) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | GSM640577(GSM640577) small RNA in the liver with paternal control . (liver) | mjTestesWT2() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................GTAAAGGCTGGGCTGAGAcgtg............................................................................................................................ | 22 | CGTG | 57.00 | 0.00 | 7.00 | 5.00 | 4.00 | - | 3.00 | - | 3.00 | 6.00 | - | 6.00 | 3.00 | 2.00 | 2.00 | 1.00 | 1.00 | 1.00 | 2.00 | - | - | 1.00 | 1.00 | - | 1.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcgt............................................................................................................................. | 21 | CGT | 57.00 | 0.00 | 8.00 | 9.00 | 8.00 | 5.00 | 7.00 | 7.00 | 3.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcg.............................................................................................................................. | 20 | CG | 11.00 | 0.00 | 1.00 | - | - | 3.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcgtt............................................................................................................................ | 22 | CGTT | 8.00 | 0.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGGgcga.......................................................................................................................... | 22 | GCGA | 6.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGGgcgt.......................................................................................................................... | 22 | GCGT | 5.00 | 2.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcga............................................................................................................................. | 21 | CGA | 4.00 | 0.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGG.............................................................................................................................. | 18 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAagt............................................................................................................................. | 21 | AGT | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcgag............................................................................................................................ | 22 | CGAG | 2.00 | 0.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAGGt............................................................................................................................. | 21 | T | 2.00 | 0.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcgc............................................................................................................................. | 21 | CGC | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAaacg.............................................................................................................................. | 20 | AACG | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAGGtggc.......................................................................................................................... | 24 | TGGC | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGtcgt............................................................................................................................. | 21 | TCGT | 1.14 | 0.43 | 0.43 | - | 0.29 | - | 0.14 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGGg............................................................................................................................. | 19 | G | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcat............................................................................................................................. | 21 | CAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAGGtg............................................................................................................................ | 22 | TG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................GCTGGGCTGAGAGGACg........................................................................................................................... | 17 | G | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAagtg............................................................................................................................ | 22 | AGTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGGgaga.......................................................................................................................... | 22 | GAGA | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAGac............................................................................................................................. | 21 | AC | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGGgggt.......................................................................................................................... | 22 | GGGT | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcgga............................................................................................................................ | 22 | CGGA | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAtgt............................................................................................................................. | 21 | TGT | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAagat............................................................................................................................ | 22 | AGAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAt................................................................................................................................. | 17 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAtttg............................................................................................................................ | 22 | TTTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAcca............................................................................................................................. | 21 | CCA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGGgcat.......................................................................................................................... | 22 | GCAT | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAagtt............................................................................................................................ | 22 | AGTT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGGgctt.......................................................................................................................... | 22 | GCTT | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAGGgcta.......................................................................................................................... | 22 | GCTA | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................AAAGGCTGGGCTGAGAcgg............................................................................................................................. | 19 | CGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................GCTGGGCTGAGAGGAtga.......................................................................................................................... | 18 | TGA | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAc................................................................................................................................. | 17 | C | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAGGgcga.......................................................................................................................... | 24 | GCGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................TGGGCTGAGAGGACActc........................................................................................................................ | 18 | CTC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAct.............................................................................................................................. | 20 | CT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AAGGCTGGGCTGAGAGGccg........................................................................................................................... | 20 | CCG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAc............................................................................................................................... | 19 | C | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGAGGtaa........................................................................................................................... | 23 | TAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AAGGCTGGGCTGAGAGGgcga.......................................................................................................................... | 21 | GCGA | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGccgt............................................................................................................................. | 21 | CCGT | 0.71 | 0.43 | 0.14 | 0.14 | 0.14 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................ATGGACCCAGTAGATA................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ............................................................GTAAAGGCTGGGCTGAG................................................................................................................................. | 17 | 7 | 0.43 | 0.43 | 0.14 | 0.14 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGACCCAGTAGATA................................................................................................. | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| ............................................................GTAAAGGCTGGGCTGAGgctt............................................................................................................................. | 21 | GCTT | 0.14 | 0.43 | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGcggt............................................................................................................................. | 21 | CGGT | 0.14 | 0.43 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGtcat............................................................................................................................. | 21 | TCAT | 0.14 | 0.43 | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGgtgt............................................................................................................................. | 21 | GTGT | 0.14 | 0.43 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGtcgg............................................................................................................................. | 21 | TCGG | 0.14 | 0.43 | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGcaat............................................................................................................................. | 21 | CAAT | 0.14 | 0.43 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGcca.............................................................................................................................. | 20 | CCA | 0.14 | 0.43 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GTAAAGGCTGGGCTGAGgcgt............................................................................................................................. | 21 | GCGT | 0.14 | 0.43 | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTTGGCATCATGTCACTTCAGGTTAGATGTACATGCTCTTGATGTACAGGGCATCATTTGGTAAAGGCTGGGCTGAGAGGACATCTTTGTTCCATGGACCCAGTAGATAGGCAGCTTCTGCTTCCAGGAACAACTTGTCTGTTTTTCAGTCTATCTACTTCCTAAAATTTCAATTTGCTCACATAGCACTTAAGAGAAGAGCTCTC ...........................................................(((....)))((((((((((((((...(((((((.((((...((((((.((....)))))))))))))))))))..)))))...)))))))))...................................................... ........................................................57.................................................................................................156................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | mjLiverWT1() Liver Data. (liver) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR095855BC6(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR059765(GSM562827) DN3_control. (thymus) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | mjTestesWT4() Testes Data. (testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR206940(GSM723281) other. (brain) | SRR073954(GSM629280) total RNA. (blood) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR037937(GSM510475) 293cand2. (cell line) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | GSM640577(GSM640577) small RNA in the liver with paternal control . (liver) | mjTestesWT2() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................ggcGTCTGTTTTTCAGTCT...................................................... | 19 | ggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................cccGTCTGTTTTTCAGTCT...................................................... | 19 | ccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................tgcGTCTGTTTTTCAGTCT...................................................... | 19 | tgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................caGCAGCTTCTGCTTCCAG............................................................................... | 19 | ca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................acgGTCTGTTTTTCAGTCT...................................................... | 19 | acg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................tttaTATCTACTTCCTAAA........................................ | 19 | ttta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................taCTATCTACTTCCTAA......................................... | 17 | ta | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |