| (1) AGO2.ip | (9) B-CELL | (5) BRAIN | (4) EMBRYO | (11) ESC | (4) FIBROBLAST | (4) LIVER | (3) LYMPH | (4) OTHER | (3) OTHER.mut | (1) PIWI.ip | (5) SPLEEN | (7) TESTES | (3) THYMUS |
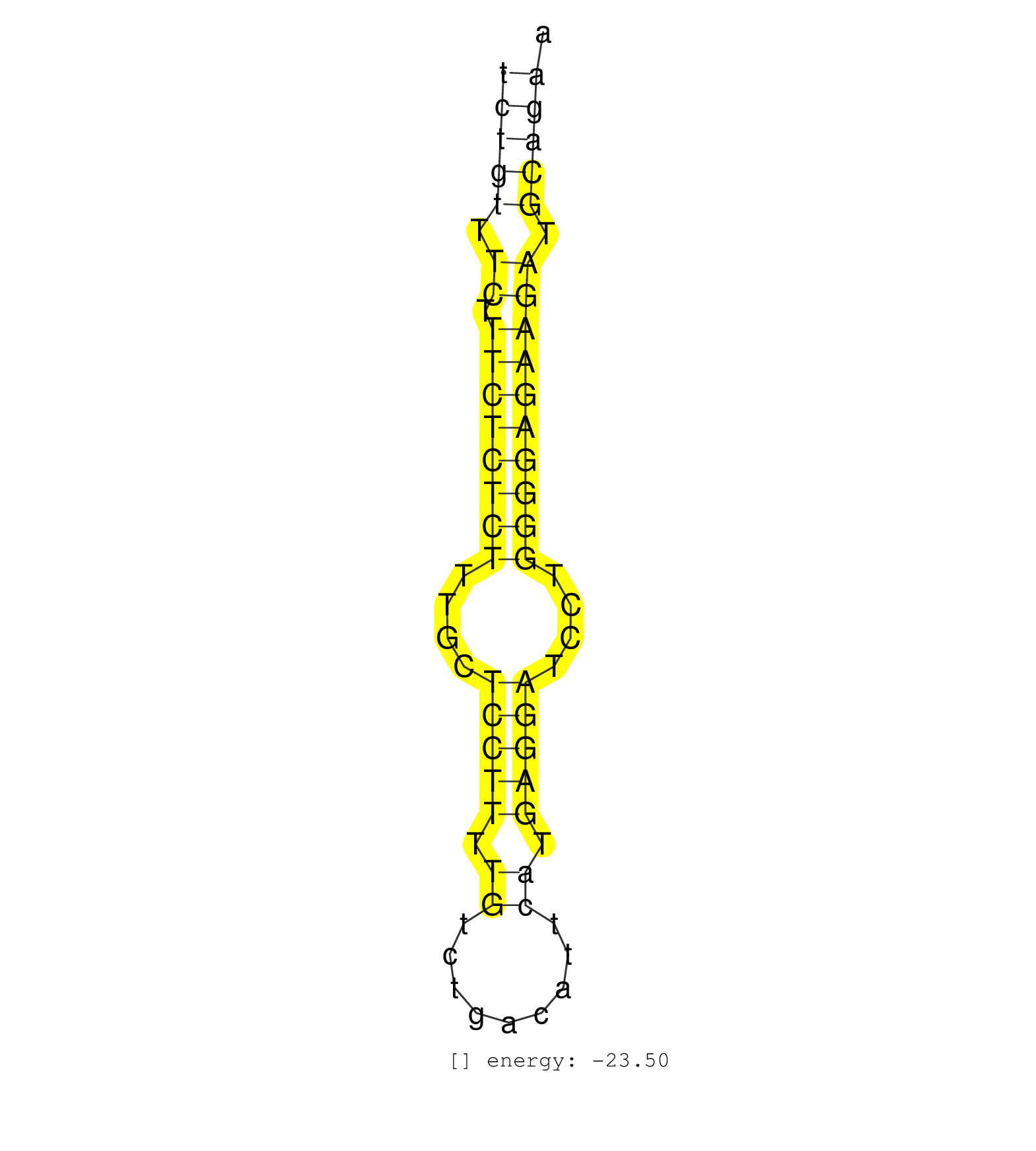
| GACGAGTTACTGTTACAGTAGAAGTAAGAGCATTTTTATTGAAATTAATGCCTGAATTCTGTTTCTTTCTCTCTTTGCTCCTTTTGTCTGACATTCATGAGGATCCTGGGGAGAAGATGCAGAATTCCCTGGAAATTGACTTACAGATCGTTGTGGGTGCTAGGAACTGGACTCGGGCTCTC ..................................................(((((.((.((((((((....(((((.((.........)).)))))....)))))))))).)))))................................................... .........................................................58................................................................124........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | mjTestesWT4() Testes Data. (testes) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR059765(GSM562827) DN3_control. (thymus) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | GSM475281(GSM475281) total RNA. (testes) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | mjLiverKO1() Liver Data. (Zcchc11 liver) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................TGAGGATCCTGGGGAGAAGATGC.............................................................. | 23 | 1 | 47.00 | 47.00 | 10.00 | 4.00 | 5.00 | 5.00 | - | 1.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | 3.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGATG............................................................... | 22 | 1 | 21.00 | 21.00 | 4.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGATGt.............................................................. | 23 | T | 7.00 | 21.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGAT................................................................ | 21 | 1 | 7.00 | 7.00 | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGATGa.............................................................. | 23 | A | 5.00 | 21.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGA................................................................. | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................TTCTTTCTCTCTTTGCTCCTTTTG................................................................................................ | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGATGCAa............................................................ | 25 | A | 3.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGAggc.............................................................. | 23 | GGC | 3.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TTCTTTCTCTCTTTGCTCCTTTTGT............................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...GAGTTACTGTTACAGTAG................................................................................................................................................................. | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGAa................................................................ | 21 | A | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAtgat................................................................ | 21 | TGAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...GAGTTACTGTTACAGTAGAAGT............................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TTCTTTCTCTCTTTGCTCCTTT.................................................................................................. | 22 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ......................................................................................................ATCCTGGGGAGAAGAcctc............................................................. | 19 | CCTC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGGAAATTGACTTACAGATCGTTGTG........................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....GTTACTGTTACAGTAGAAGTAAGAGCATT.................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGACTTACAGATCGTTGTGGG......................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AGGATCCTGGGGAGAAGATGa.............................................................. | 21 | A | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGctgc.............................................................. | 23 | CTGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGATa............................................................... | 22 | A | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GATCGTTGTGGGTGCTAGGAACT.............. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................GAGGATCCTGGGGAGAAGATGC.............................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................AGAAGTAAGAGCATTTTTAT............................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGATGttt............................................................ | 25 | TTT | 1.00 | 21.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAA................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AGGATCCTGGGGAGAAGATGttt............................................................ | 23 | TTT | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGATGtaa............................................................ | 25 | TAA | 1.00 | 21.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGATCCTGGGGAGAAGATGCA............................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TCCTTTTGTCTGACAcgg...................................................................................... | 18 | CGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................CCTGGGGAGAAGATGCAG............................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GACGAGTTACTGTTACAGTAGAAGTAAGAGCATTTTTATTGAAATTAATGCCTGAATTCTGTTTCTTTCTCTCTTTGCTCCTTTTGTCTGACATTCATGAGGATCCTGGGGAGAAGATGCAGAATTCCCTGGAAATTGACTTACAGATCGTTGTGGGTGCTAGGAACTGGACTCGGGCTCTC ..................................................(((((.((.((((((((....(((((.((.........)).)))))....)))))))))).)))))................................................... .........................................................58................................................................124........................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | mjTestesWT4() Testes Data. (testes) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR059765(GSM562827) DN3_control. (thymus) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | GSM475281(GSM475281) total RNA. (testes) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | mjLiverKO1() Liver Data. (Zcchc11 liver) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................TGAGGATCCTGGGGAGAAGATGC.............................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| gtgCGAGTTACTGTTACA..................................................................................................................................................................... | 18 | gtg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................cgtgCTTTGCTCCTTTTGT............................................................................................... | 19 | cgtg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................aatAATTCTGTTTCTTTC................................................................................................................. | 18 | aat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .ACGAGTTACTGTTAC...................................................................................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |