| (3) BRAIN | (7) EMBRYO | (1) ESC | (1) LIVER | (1) OTHER | (1) SPLEEN | (1) TESTES |
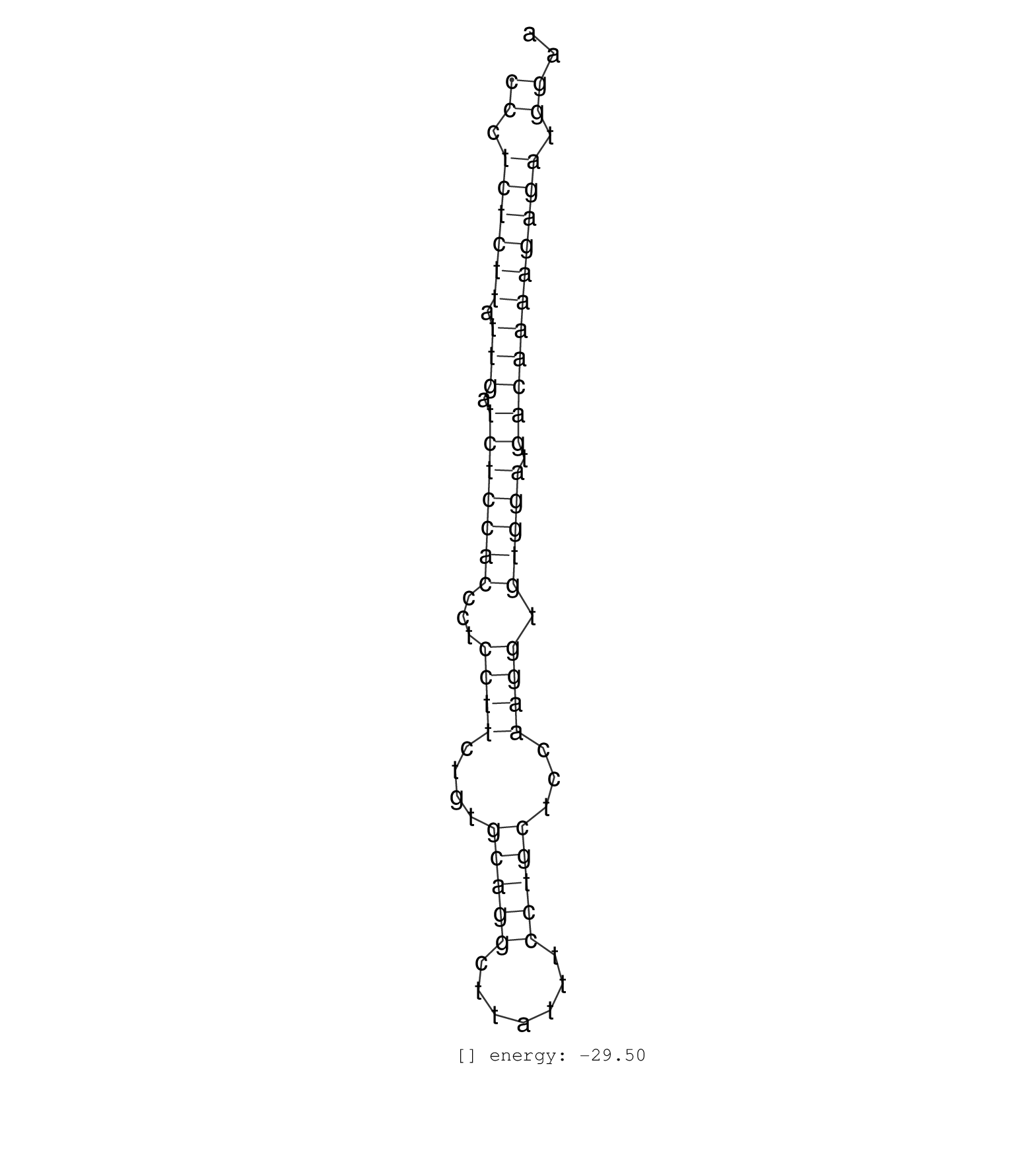
| GTCTAAATAAATGTTGGTATTGAGACAAGGCAAATATTGCAGCCACCTATCCCTCTCTTATTGATCTCCACCCTCCTTCTGTGCAGGCTTATTTCCTGCTCCAAGGTGTGGATGACAAAAGAGATGGAAGTATGGGGGGGCGGGAGGGCACACAGAGGCTTTCATAAACCCTCTCTGCT ..................................................((.((((((.(((.(((((((...((((....(((((.......)))))...)))).))))).))))))))))).)).................................................... ..................................................51............................................................................129................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | GSM640582(GSM640582) small RNA in the liver with paternal control . (liver) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR073954(GSM629280) total RNA. (blood) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................TCTCCACCCTCCTTCTGct................................................................................................ | 19 | CT | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TCTCCACCCTCCTTCgatt................................................................................................ | 19 | GATT | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................TCTCCACCCTCCTTCTact................................................................................................ | 19 | ACT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................TCTCCACCCTCCTTCgtct................................................................................................ | 19 | GTCT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TCTCCACCCTCCTTCggtt................................................................................................ | 19 | GGTT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGGGCGGGAGGGCAttt.......................... | 18 | TTT | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGGGCGGGAGGGCAaaaa......................... | 19 | AAAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................TCTCCACCCTCCTTCgccc................................................................................................ | 19 | GCCC | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................ACACAGAGGCTTTCAaat............ | 18 | AAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................TCTCCACCCTCCTTCTacc................................................................................................ | 19 | ACC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................GGGGGGGCGGGAGGGgcgg........................... | 19 | GCGG | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........AATGTTGGTATTGAGc.......................................................................................................................................................... | 16 | C | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TCTCCACCCTCCTTCgtgc................................................................................................ | 19 | GTGC | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................TCTCCACCCTCCTTCTGTc................................................................................................ | 19 | C | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TCTCCACCCTCCTTCggtc................................................................................................ | 19 | GGTC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| GTCTAAATAAATGTTGGTATTGAGACAAGGCAAATATTGCAGCCACCTATCCCTCTCTTATTGATCTCCACCCTCCTTCTGTGCAGGCTTATTTCCTGCTCCAAGGTGTGGATGACAAAAGAGATGGAAGTATGGGGGGGCGGGAGGGCACACAGAGGCTTTCATAAACCCTCTCTGCT ..................................................((.((((((.(((.(((((((...((((....(((((.......)))))...)))).))))).))))))))))).)).................................................... ..................................................51............................................................................129................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | GSM640582(GSM640582) small RNA in the liver with paternal control . (liver) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR073954(GSM629280) total RNA. (blood) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................agcgTGATCTCCACCCTCC....................................................................................................... | 19 | agcg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................gacGGGGGGCGGGAGGGC.............................. | 18 | gac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................gcCAGAGGCTTTCATAA............ | 17 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |