| (1) AGO2.ip | (1) B-CELL | (16) BREAST | (7) CELL-LINE | (1) CERVIX | (2) LIVER | (2) OTHER | (2) PLACENTA |
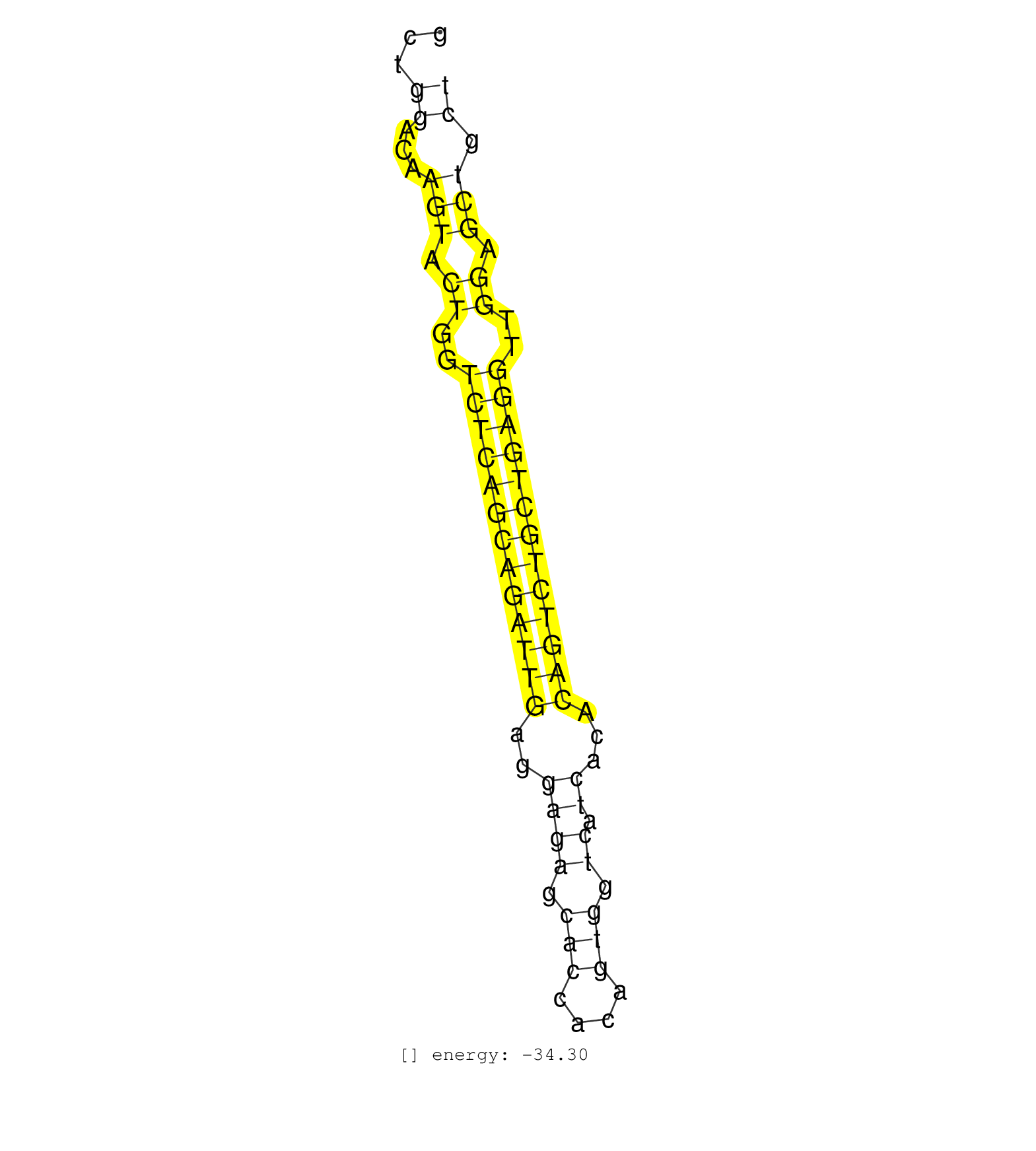
| GATCATGGCAGACATTCGGCCCAATACGATGAGCTGGCTGGGAAGAACCGAGAGAAGCTGGACAAGTACTGGTCTCAGCAGATTGAGGAGAGCACCACAGTGGTCATCACACAGTCTGCTGAGGTTGGAGCTGCTGAGATGACACTCACGGAGCTGAGACATACAGTCTAGTCCTTGGAGATCGACTTGGACTCCA .....................................................((...(((.((..(((((((((((((..((((.(((....))).)).))...)))))))))))))..)).))).)).................................................. ........................................................57............................................................................135........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR189784 | GSM532876(GSM532876) G547T. (cervix) | SRR191633(GSM715743) 85genomic small RNA (size selected RNA from t. (breast) | SRR189782 | SRR191626(GSM715736) 36genomic small RNA (size selected RNA from t. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR139199(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR191420(GSM715530) 122genomic small RNA (size selected RNA from . (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR191612(GSM715722) 65genomic small RNA (size selected RNA from t. (breast) | SRR191630(GSM715740) 70genomic small RNA (size selected RNA from t. (breast) | SRR191631(GSM715741) 75genomic small RNA (size selected RNA from t. (breast) | SRR191610(GSM715720) 195genomic small RNA (size selected RNA from . (breast) | SRR191634(GSM715744) 98genomic small RNA (size selected RNA from t. (breast) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR191625(GSM715735) 32genomic small RNA (size selected RNA from t. (breast) | SRR191623(GSM715733) 18genomic small RNA (size selected RNA from t. (breast) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | GSM359197(GSM359197) hepg2_depl_cip_tap. (cell line) | GSM359196(GSM359196) m3m7G_ip. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................ACAGTCTGCTGAGGTTGGAGC................................................................. | 21 | 2 | 7.00 | 7.00 | 0.50 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 0.50 | 0.50 | 0.50 | - | 0.50 | 0.50 | 0.50 | - | 0.50 | 0.50 | 0.50 | - | - |
| ..................................................................................................................TCTGCTGAGGTTGGAGCTGC.............................................................. | 20 | 2 | 3.50 | 3.50 | 3.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GAGCACCACAGTGGTttg......................................................................................... | 18 | TTG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CTGCTGAGGTTGGAGCTGCTG............................................................ | 21 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CTGGCTGGGAAGAACtcat................................................................................................................................................ | 19 | TCAT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................GTCTGCTGAGGTTGGctg................................................................. | 18 | CTG | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AGCTGCTGAGATGACta................................................... | 17 | TA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................ACAGTCTGCTGAGGTTGGAGt................................................................. | 21 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TCTGCTGAGGTTGGAGCTGCTGA........................................................... | 23 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ...................................................GAGAAGCTGGACAAGaacg.............................................................................................................................. | 19 | AACG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................GTCTGCTGAGGTTGGAGCTGCTGAGA......................................................... | 26 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................AGTCTGCTGAGGTTGGAtct................................................................ | 20 | TCT | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................CTCAGCAGATTGAGGgc.......................................................................................................... | 17 | GC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................AGTCTGCTGAGGTTGGAGCTGCTG............................................................ | 24 | 2 | 1.00 | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................ACAGTCTGCTGAGGTTGagc.................................................................. | 20 | AGC | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCTGAGGTTGGAGCTGCTGA........................................................... | 21 | 2 | 1.00 | 1.00 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCTGAGGTTGGAGCTGC.............................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGCTGAGGTTGGAGCTGCTGAG.......................................................... | 22 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CAGTCTGCTGAGGTTGGAGC................................................................. | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................GTCTGCTGAGGTTGGAG.................................................................. | 17 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TCTGCTGAGGTTGGAGCTGCTG............................................................ | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................GTCTGCTGAGGTTGGAGCTGCTGA........................................................... | 24 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CAGTCTGCTGAGGTTGGAGCTGCT............................................................. | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGAGGTTGGAGCTGCTGA........................................................... | 18 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGAGGTTGGAGCTGCTGAa.......................................................... | 19 | A | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CTGGTCTCAGCAGATTGAGGAGAGCACCACAGTGGT............................................................................................ | 36 | 8 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | 0.12 |
| ..................................................................TACTGGTCTCAGCAGATTGAGGAGAGCACCACAGTG.............................................................................................. | 36 | 8 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | 0.12 |
| GATCATGGCAGACATTCGGCCCAATACGATGAGCTGGCTGGGAAGAACCGAGAGAAGCTGGACAAGTACTGGTCTCAGCAGATTGAGGAGAGCACCACAGTGGTCATCACACAGTCTGCTGAGGTTGGAGCTGCTGAGATGACACTCACGGAGCTGAGACATACAGTCTAGTCCTTGGAGATCGACTTGGACTCCA .....................................................((...(((.((..(((((((((((((..((((.(((....))).)).))...)))))))))))))..)).))).)).................................................. ........................................................57............................................................................135........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR189784 | GSM532876(GSM532876) G547T. (cervix) | SRR191633(GSM715743) 85genomic small RNA (size selected RNA from t. (breast) | SRR189782 | SRR191626(GSM715736) 36genomic small RNA (size selected RNA from t. (breast) | TAX577741(Rovira) total RNA. (breast) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR139199(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR191420(GSM715530) 122genomic small RNA (size selected RNA from . (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR191612(GSM715722) 65genomic small RNA (size selected RNA from t. (breast) | SRR191630(GSM715740) 70genomic small RNA (size selected RNA from t. (breast) | SRR191631(GSM715741) 75genomic small RNA (size selected RNA from t. (breast) | SRR191610(GSM715720) 195genomic small RNA (size selected RNA from . (breast) | SRR191634(GSM715744) 98genomic small RNA (size selected RNA from t. (breast) | SRR191635(GSM715745) 9genomic small RNA (size selected RNA from to. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR191625(GSM715735) 32genomic small RNA (size selected RNA from t. (breast) | SRR191623(GSM715733) 18genomic small RNA (size selected RNA from t. (breast) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | GSM359197(GSM359197) hepg2_depl_cip_tap. (cell line) | GSM359196(GSM359196) m3m7G_ip. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................gttaTCACGGAGCTGAGACA................................... | 20 | gtta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................tagaAGGTTGGAGCTGCTG............................................................ | 19 | taga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |