| (1) AGO2.ip | (3) BREAST | (7) CELL-LINE | (2) HEART | (1) LIVER | (2) OTHER | (2) PLACENTA | (1) RRP40.ip | (1) SPLEEN | (1) TESTES |
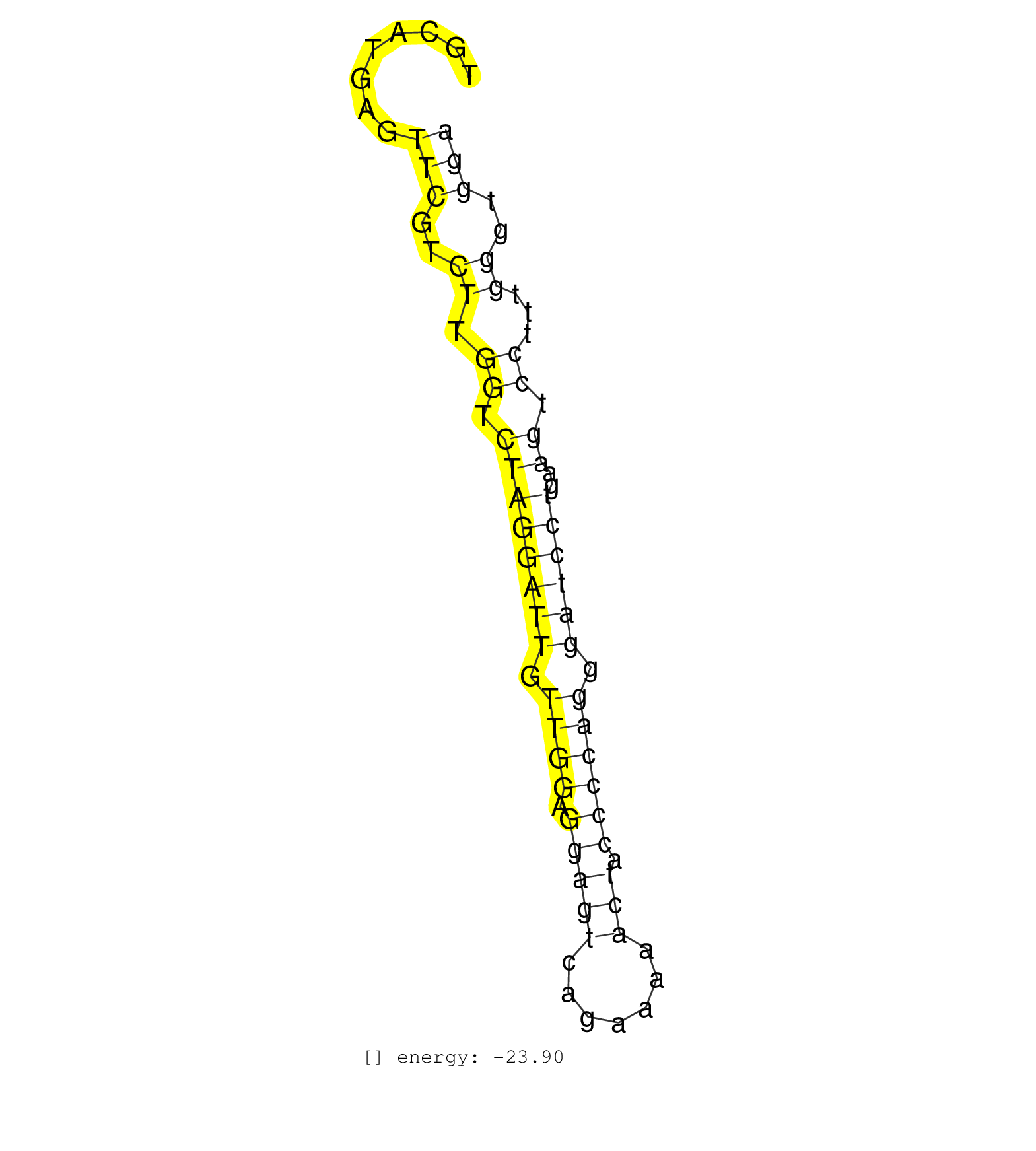
| CCAGACAACTCAGCGGCTACACGCTGACTTCCTGTGATGCCTTGAAAAAGTGCATGAGTTCGTCTTGGTCTAGGATTGTTGGAGGAGTCAGAAAAACTACCCCAGGGATCCTGAAGTCCTTTGGGTGGAAAGGGGGAGGCCCCCATGTCTGTCTGGCTCTCAGAAAATGGGAAAGAAGG ..........................................................(((..((.((.((((((((.((((.(((((.......))).)))))).))))))..)).))...))..))).................................................. ..................................................51............................................................................129................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR139199(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR037937(GSM510475) 293cand2. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189784 | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR139205(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR191633(GSM715743) 85genomic small RNA (size selected RNA from t. (breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR189782 | TAX577579(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................GGAGGAGTCAGAAAAgtt................................................................................. | 18 | GTT | 4.00 | 0.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGGTCTAGGATTGTTGGAGGtgg........................................................................................... | 23 | TGG | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................GGAGGAGTCAGAAAAgt.................................................................................. | 17 | GT | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................TCTAGGATTGTTGGAGGAGT........................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................GGTCTAGGATTGTTGGAGGAa............................................................................................ | 21 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....CAACTCAGCGGCTACAta............................................................................................................................................................ | 18 | TA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................AAAAACTACCCCAGGGtac..................................................................... | 19 | TAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................AAGGGGGAGGCCCCCggct............................... | 19 | GGCT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................GGAAAGGGGGAGGCCgg.................................... | 17 | GG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................GGAGTCAGAAAAACTACCataa.......................................................................... | 22 | ATAA | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....ACAACTCAGCGGCTACAt............................................................................................................................................................. | 18 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................TGGTCTAGGATTGTTGGAGGAa............................................................................................ | 22 | A | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GTGCATGAGTTCGTCTTGGTCTAGGATTGTTGGAG............................................................................................... | 35 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGGTCTAGGATTGTTGGAGGgt............................................................................................ | 22 | GT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AAGGGGGAGGCCCCCggcg............................... | 19 | GGCG | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGGTCTAGGATTGTTGGAGGAag........................................................................................... | 23 | AG | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .CAGACAACTCAGCGGC.................................................................................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| CCAGACAACTCAGCGGCTACACGCTGACTTCCTGTGATGCCTTGAAAAAGTGCATGAGTTCGTCTTGGTCTAGGATTGTTGGAGGAGTCAGAAAAACTACCCCAGGGATCCTGAAGTCCTTTGGGTGGAAAGGGGGAGGCCCCCATGTCTGTCTGGCTCTCAGAAAATGGGAAAGAAGG ..........................................................(((..((.((.((((((((.((((.(((((.......))).)))))).))))))..)).))...))..))).................................................. ..................................................51............................................................................129................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR139199(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR037937(GSM510475) 293cand2. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189784 | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR139205(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR191633(GSM715743) 85genomic small RNA (size selected RNA from t. (breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR189782 | TAX577579(Rovira) total RNA. (breast) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................CAGGGATCCTGAAGT.............................................................. | 15 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |