| (1) AGO2.ip | (2) B-CELL | (13) BREAST | (27) CELL-LINE | (4) CERVIX | (1) FIBROBLAST | (1) HEART | (2) HELA | (1) LUNG | (4) OTHER | (2) OVARY | (1) PLACENTA | (1) RRP40.ip | (10) SKIN | (1) XRN.ip |
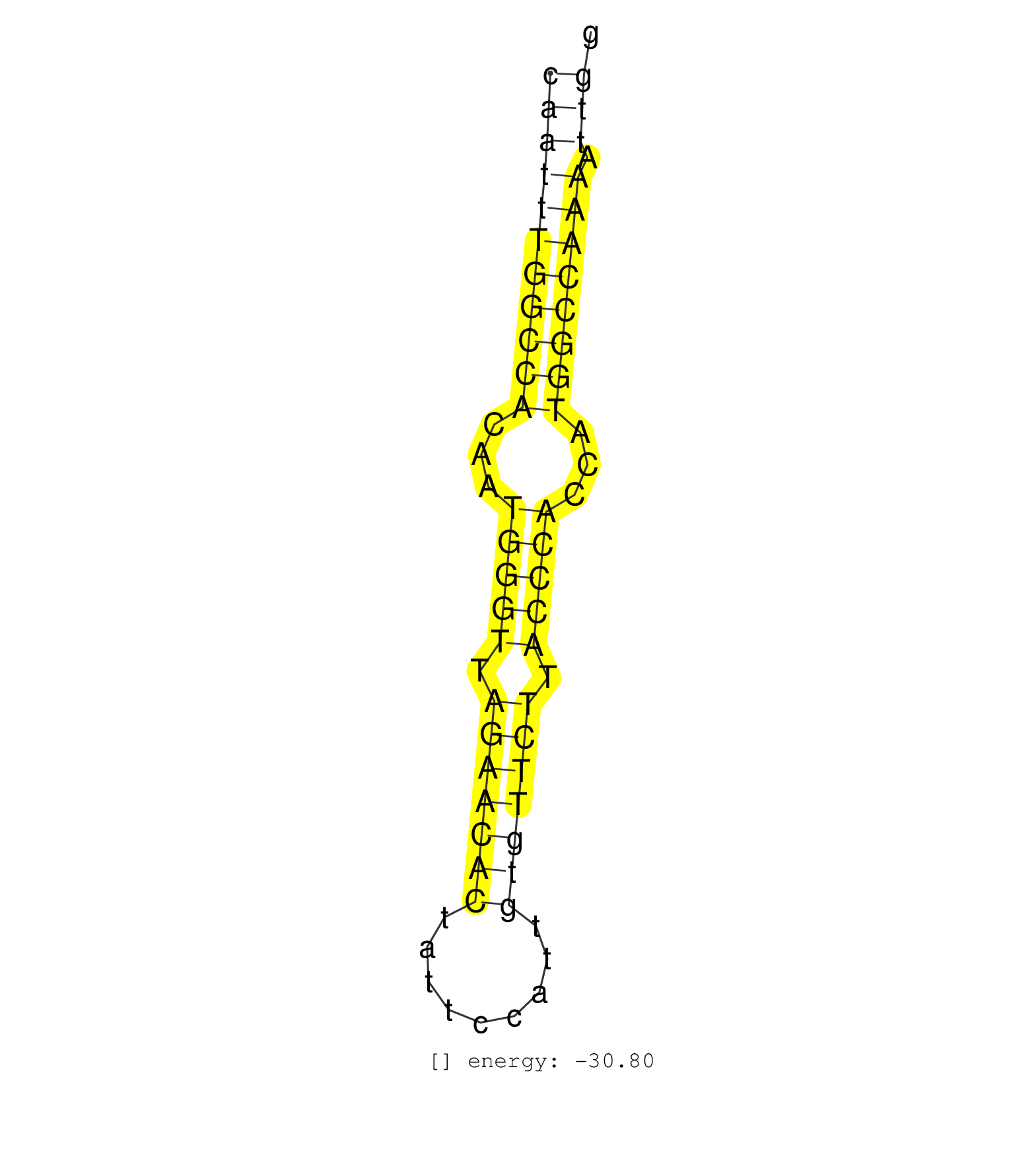
| AATTGAACAACTATTTAAATAAAAAAGCACCTTCATGAGAACCGAAAATCAGCTTAGGTACCAATTTGGCCACAATGGGTTAGAACACTATTCCATTGTGTTCTTACCCACCATGGCCAAAATTGGGCCTAAGGGGTCCCAATTCCCGGCCTCAGCTCTTGGATGGAATTTCTAGACCTGCCC ..................................................(((((((((((...(((((.(((((((.........))))))).)))))...)))))))).)))................................................... .............................................................62..............................................................126....................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR189783 | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR040010(GSM532895) G529N. (cervix) | GSM416733(GSM416733) HEK293. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189786 | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR037937(GSM510475) 293cand2. (cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR189782 | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR040018(GSM532903) G701N. (cervix) | SRR191420(GSM715530) 122genomic small RNA (size selected RNA from . (breast) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR191462(GSM715572) 1genomic small RNA (size selected RNA from to. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR139191(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR139190(SRX050652) FLASHPage purified small RNA (~15-40nt) from . (lung) | TAX577740(Rovira) total RNA. (breast) | SRR189785 | SRR191535(GSM715645) 181genomic small RNA (size selected RNA from . (breast) | SRR191406(GSM715516) 67genomic small RNA (size selected RNA from t. (breast) | SRR191495(GSM715605) 159genomic small RNA (size selected RNA from . (breast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR191518(GSM715628) 83genomic small RNA (size selected RNA from t. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR139193(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR040024(GSM532909) G613N. (cervix) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR191601(GSM715711) 58genomic small RNA (size selected RNA from t. (breast) | SRR191563(GSM715673) 83genomic small RNA (size selected RNA from t. (breast) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................TGGCCACAATGGGTTAGAACAC............................................................................................... | 22 | 1 | 47.00 | 47.00 | 9.00 | - | 5.00 | 10.00 | 3.00 | - | - | 6.00 | 3.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ..................................................................TGGCCACAATGGGTTAGAAC................................................................................................. | 20 | 1 | 27.00 | 27.00 | 2.00 | 4.00 | 8.00 | - | 1.00 | - | - | - | 3.00 | - | 5.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................TGGCCACAATGGGTTAGAACA................................................................................................ | 21 | 1 | 26.00 | 26.00 | 6.00 | - | - | - | 2.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ....................................................................................................TTCTTACCCACCATGGCCAAAA............................................................. | 22 | 1 | 25.00 | 25.00 | 7.00 | - | - | - | 2.00 | 1.00 | 3.00 | - | - | - | - | 3.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - |
| ....................................................................................................TTCTTACCCACCATGGCCAAA.............................................................. | 21 | 1 | 15.00 | 15.00 | - | 8.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TTGGCCACAATGGGTTAGAAC................................................................................................. | 21 | 1 | 13.00 | 13.00 | 5.00 | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................AAATCAGCTTAGGTACCAATT..................................................................................................................... | 21 | 1 | 12.00 | 12.00 | - | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGCCAA............................................................... | 20 | 1 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................TGGCCACAATGGGTTAGAACACT.............................................................................................. | 23 | 1 | 8.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGCCAAAAT............................................................ | 23 | 1 | 7.00 | 7.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGGCCACAATGGGTTAGAACACc.............................................................................................. | 23 | C | 6.00 | 47.00 | 2.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGGCCACAATGGGTTAG.................................................................................................... | 17 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GGCCACAATGGGTTAGAACA................................................................................................ | 20 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TCTTACCCACCATGGCCAAAA............................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGCCAAt.............................................................. | 21 | T | 3.00 | 9.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGCCAga.............................................................. | 21 | GA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CCACAATGGGTTAGAACA................................................................................................ | 18 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CCACAATGGGTTAGAACAC............................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGCCca............................................................... | 20 | CA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGGCCACAATGGGTTAGAAa................................................................................................. | 20 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGC.................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGCCAt............................................................... | 20 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGCCAAAAa............................................................ | 23 | A | 1.00 | 25.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TTGGCCACAATGGGTTAGAACA................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGGCCACAATGGGTTAGAACACa.............................................................................................. | 23 | A | 1.00 | 47.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................TTGGCCACAATGGGTTAGAACcgcc............................................................................................. | 25 | CGCC | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................ATCAGCTTAGGTACCAATT..................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGGac................................................................. | 18 | AC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGGCCACAATGGGTTAGAAaa................................................................................................ | 21 | AA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGGCCACAATGGGTTAGAACACTc............................................................................................. | 24 | C | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGGCCACAATGGGTTAGAAtacc.............................................................................................. | 23 | TACC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATGctaa................................................................ | 19 | CTAA | 0.29 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTCTTACCCACCATG.................................................................... | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AATTGAACAACTATTTAAATAAAAAAGCACCTTCATGAGAACCGAAAATCAGCTTAGGTACCAATTTGGCCACAATGGGTTAGAACACTATTCCATTGTGTTCTTACCCACCATGGCCAAAATTGGGCCTAAGGGGTCCCAATTCCCGGCCTCAGCTCTTGGATGGAATTTCTAGACCTGCCC ..................................................(((((((((((...(((((.(((((((.........))))))).)))))...)))))))).)))................................................... .............................................................62..............................................................126....................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR189783 | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037931(GSM510469) 293GFP. (cell line) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR040010(GSM532895) G529N. (cervix) | GSM416733(GSM416733) HEK293. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189786 | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR037937(GSM510475) 293cand2. (cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR189782 | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR040018(GSM532903) G701N. (cervix) | SRR191420(GSM715530) 122genomic small RNA (size selected RNA from . (breast) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR029124(GSM416753) HeLa. (hela) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR191462(GSM715572) 1genomic small RNA (size selected RNA from to. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR139191(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR139190(SRX050652) FLASHPage purified small RNA (~15-40nt) from . (lung) | TAX577740(Rovira) total RNA. (breast) | SRR189785 | SRR191535(GSM715645) 181genomic small RNA (size selected RNA from . (breast) | SRR191406(GSM715516) 67genomic small RNA (size selected RNA from t. (breast) | SRR191495(GSM715605) 159genomic small RNA (size selected RNA from . (breast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR191518(GSM715628) 83genomic small RNA (size selected RNA from t. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR139193(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR040024(GSM532909) G613N. (cervix) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR191601(GSM715711) 58genomic small RNA (size selected RNA from t. (breast) | SRR191563(GSM715673) 83genomic small RNA (size selected RNA from t. (breast) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........acagTTTAAATAAAAAAGC........................................................................................................................................................... | 19 | acag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |