| (1) AGO1.ip | (1) AGO2.ip | (15) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (1) KIDNEY | (1) LIVER | (1) OTHER | (1) OVARY | (1) PLACENTA | (2) SKIN | (1) THYMUS | (1) XRN.ip |
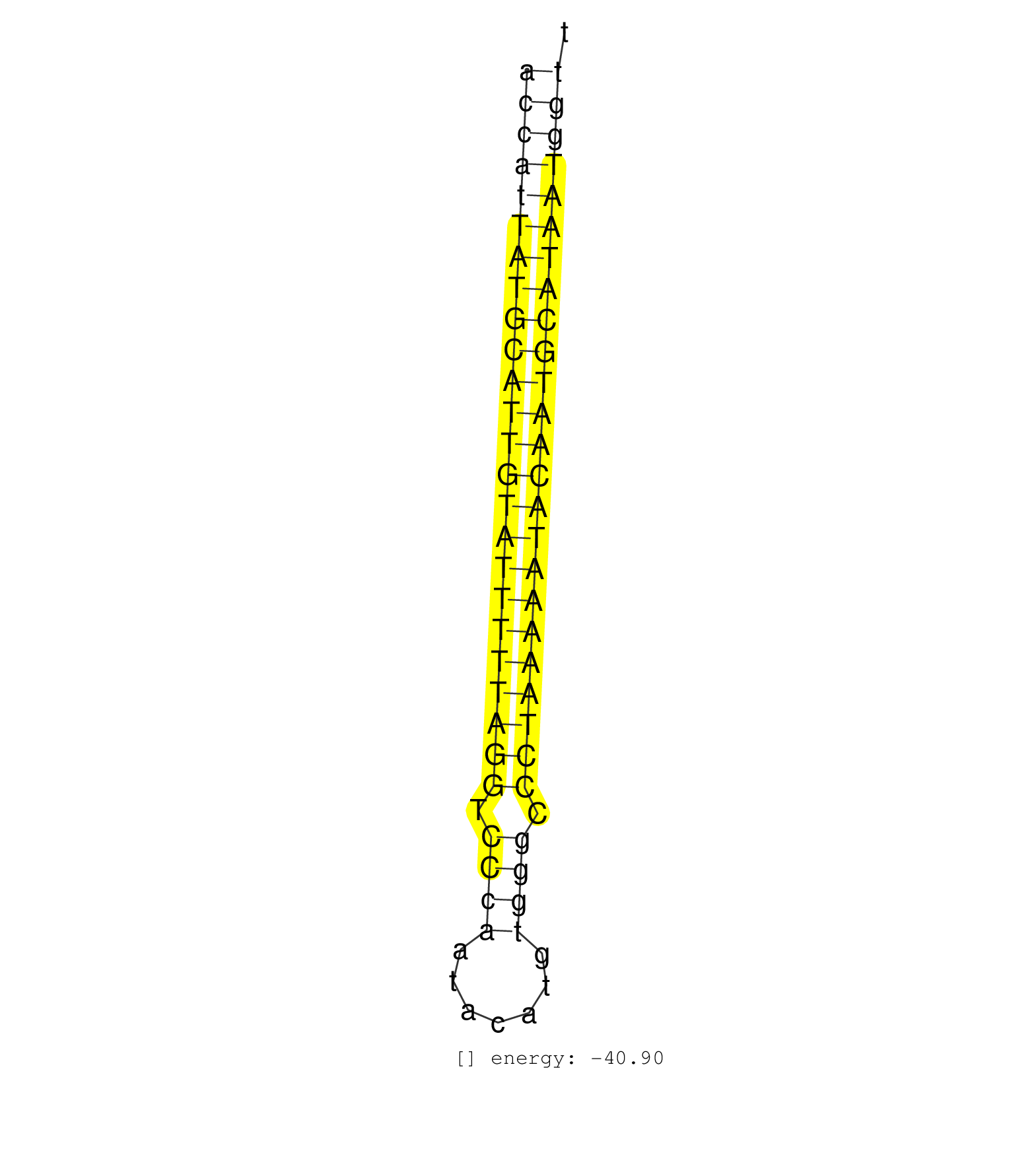
| AATTCAAAGAAAAGCAATTATCTAACAAAGTCAAATTCTTATTAAAAGACATGGGGTAAAACCATTATGCATTGTATTTTTAGGTCCCAATACATGTGGGCCCTAAAAATACAATGCATAATGGTTTTTCACTCTTTATCTTCTTATTTTATGTAAATGAAAAAATAGTATTGTCAATTTTTTTCTGCAAATACCAC ..................................................((((((((((((((((((((((((.((((.......)))).))))))))))))))))))))))))................................................... ............................................................61...............................................................126..................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189782 | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189783 | SRR189784 | SRR037937(GSM510475) 293cand2. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR189785 | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR040037(GSM532922) G243T. (cervix) | SRR189787 | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR189786 | SRR139217(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................TATGCATTGTATTTTTAGGTCC.............................................................................................................. | 22 | 1 | 50.00 | 50.00 | 2.00 | 11.00 | 12.00 | 9.00 | 1.00 | 4.00 | - | - | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .................................................................TATGCATTGTATTTTTAG.................................................................................................................. | 18 | 2 | 40.00 | 40.00 | 16.00 | - | - | - | - | - | 2.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CCCTAAAAATACAATGCATAAT........................................................................... | 22 | 1 | 11.00 | 11.00 | - | 6.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................TATGCATTGTATTTTTAGGTCa.............................................................................................................. | 22 | A | 11.00 | 5.00 | - | - | - | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TTATGCATTGTATTTTTAG.................................................................................................................. | 19 | 2 | 7.00 | 7.00 | 3.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TTATGCATTGTATTTTTAGGTC............................................................................................................... | 22 | 1 | 6.00 | 6.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TATGCATTGTATTTTTAGGTC............................................................................................................... | 21 | 1 | 5.00 | 5.00 | - | - | - | - | - | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGCATTGTATTTTTAGGTCCC............................................................................................................. | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GCCCTAAAAATACAATGCATAA............................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CCCTAAAAATACAATGCATAct........................................................................... | 22 | CT | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGCATTGTATTTTTAGGTCC.............................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................ATTATGCATTGTATTTTTAGGTCCCAATACA....................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GTAAATGAAAAAATAcgtg.......................... | 19 | CGTG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGCATTGTATTTTTAGGTCCCgt........................................................................................................... | 24 | GT | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TATGCATTGTATTTTTAGGTaaa............................................................................................................. | 23 | AAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................GGCCCTAAAAATACAcct................................................................................. | 18 | CCT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TATGCATTGTATTTTTAGGccc.............................................................................................................. | 22 | CCC | 1.00 | 0.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................AAAGACATGGGGTAAAtatt..................................................................................................................................... | 20 | TATT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................ATGCATTGTATTTTTAGGTCCCt............................................................................................................ | 23 | T | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GCCCTAAAAATACAATGCATAAT........................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TATTGTCAATTTTTTattt.......... | 19 | ATTT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GTAAATGAAAAAATAcacg.......................... | 19 | CACG | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ATCTTCTTATTTTATcag.......................................... | 18 | CAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................TATGCATTGTATTTTTAGGacc.............................................................................................................. | 22 | ACC | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| .................................................................TATGCATTGTATTTTTAGtgac.............................................................................................................. | 22 | TGAC | 0.50 | 40.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................ATGCATTGTATTTTTAG.................................................................................................................. | 17 | 5 | 0.40 | 0.40 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AATTCAAAGAAAAGCAATTATCTAACAAAGTCAAATTCTTATTAAAAGACATGGGGTAAAACCATTATGCATTGTATTTTTAGGTCCCAATACATGTGGGCCCTAAAAATACAATGCATAATGGTTTTTCACTCTTTATCTTCTTATTTTATGTAAATGAAAAAATAGTATTGTCAATTTTTTTCTGCAAATACCAC ..................................................((((((((((((((((((((((((.((((.......)))).))))))))))))))))))))))))................................................... ............................................................61...............................................................126..................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR189782 | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR189783 | SRR189784 | SRR037937(GSM510475) 293cand2. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR189785 | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR040037(GSM532922) G243T. (cervix) | SRR189787 | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR139201(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR189786 | SRR139217(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................CTAAAAATACAATGCATA............................................................................. | 18 | 2 | 40.00 | 40.00 | 16.00 | - | - | - | - | - | 2.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTAAAAATACAATGCATAA............................................................................ | 19 | 2 | 7.00 | 7.00 | 3.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................caatATGAAAAAATAGTAT.......................... | 19 | caat | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................gggCCTAAAAATACAATGCATA............................................................................. | 22 | ggg | 1.00 | 0.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................acttGTAAAACCATTATGC............................................................................................................................... | 19 | actt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................ctcaTTTTTCACTCTTTAT.......................................................... | 19 | ctca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..tgatAAGAAAAGCAATTAT................................................................................................................................................................................ | 19 | tgat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................ggtCCTAAAAATACAATGCATA............................................................................. | 22 | ggt | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................gtcaCTAAAAATACAATGCATA............................................................................. | 22 | gtca | 0.50 | 40.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTAAAAATACAATGCAT.............................................................................. | 17 | 5 | 0.40 | 0.40 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |