| (1) AGO2.ip | (2) B-CELL | (2) BREAST | (10) CELL-LINE | (4) CERVIX | (4) HEART | (2) HELA | (1) LIVER | (2) LUNG | (2) OTHER | (1) OVARY | (8) SKIN |
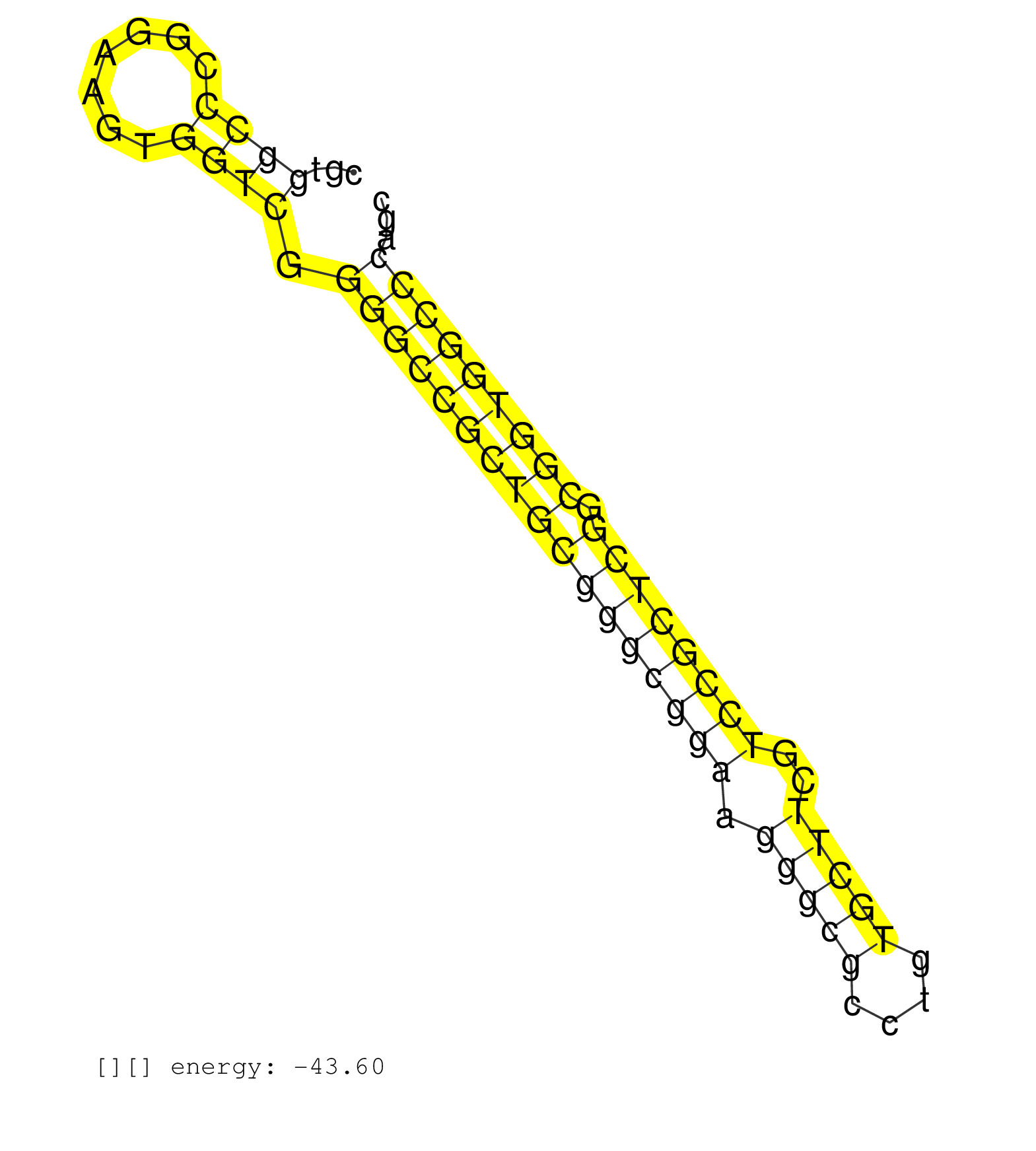
| GCGGGCTCCGAGAAGAAAGAGTGTGGCCCGGGGCGGGCGCATTAGGAGGTGTCGAGGCCGTGGCCCGGAAGTGGTCGGGGCCGCTGCGGGCGGAAGGGCGCCTGTGCTTCGTCCGCTCGGCGGTGGCCCAGCCAGGCCCGCGGGACTCAGACCAGCGGGGAGCGCGACCTCCGCCCTTGGGGCTCTCCCGCTGGG .....................................................((((.......)))).(((((((((((((((((.(((((....)))))..)))))))).)))))))))..................................................... ..........................................................59.......................................................................132............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR040016(GSM532901) G645N. (cervix) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040034(GSM532919) G001N. (cervix) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR207110(GSM721072) Nuclear RNA. (cell line) | GSM532876(GSM532876) G547T. (cervix) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR139184(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | TAX577742(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR139183(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR040019(GSM532904) G701T. (cervix) | SRR139167(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189782 | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR139192(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029125(GSM416754) U2OS. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGC............................................................... | 22 | 2 | 2.50 | 2.50 | 1.00 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - |
| ...........................................................................................................................................GCGGGACTCAGACCAGCGGGaa.................................. | 22 | AA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................GCCCGGGGCGGGCGCcgg........................................................................................................................................................ | 18 | CGG | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................AGGCCCGCGGGACTCAGA............................................ | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGCatc............................................................ | 25 | ATC | 1.00 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGCa.............................................................. | 23 | A | 1.00 | 2.50 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................CTGCGGGCGGAAGGGC................................................................................................ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GCTCGGCGGTGGCCCAttta............................................................. | 20 | TTTA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGtaa............................................................. | 24 | TAA | 1.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GGGCCGCTGCGGGCGGgggg.................................................................................................. | 20 | GGGG | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................AGTGTGGCCCGGGGCGGGCGgg.......................................................................................................................................................... | 22 | GG | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................GTGGCCCGGGGCGGGgt............................................................................................................................................................ | 17 | GT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CGGTGGCCCAGCCAGGCCCGCGGGACcc............................................... | 28 | CC | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTGCTTCGTCCGCTCGGC.......................................................................... | 18 | 2 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GCTGCGGGCGGAAGGGC................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCGGGCGGAAGGGCtag............................................................................................. | 18 | TAG | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAatt.............................................................. | 23 | ATT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGCtaat........................................................... | 26 | TAAT | 0.50 | 2.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGtt.............................................................. | 23 | TT | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGCTTCGTCCGCTCGGCGGTGGCC................................................................... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGCatt............................................................ | 25 | ATT | 0.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AGGGCGCCTGTGCTTCGTC.................................................................................. | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GAAGGGCGCCTGTGCTTCGT................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGTGCTTCGTCCGCTCGGCGG........................................................................ | 21 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTGCTTCGTCCGCTCGGCGG........................................................................ | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGt............................................................... | 22 | T | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GGGACTCAGACCAGCGGG.................................... | 18 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................AAGGGCGCCTGTGCTTCGTC.................................................................................. | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AGCCAGGCCCGCGGGAC................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CGCGGGACTCAGACCAGCGGG.................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................AGGCCCGCGGGACTCAGACC.......................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ..........................................................................................................................................CGCGGGACTCAGACCAGCGG..................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ........................................................................................................TGCTTCGTCCGCTCGGCGGTGG..................................................................... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCCGGAAGTGGTCGGGGCCGCTGC............................................................................................................ | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAGa............................................................... | 22 | A | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ..............................................................................................................GTCCGCTCGGCGGTGGCCCAG................................................................ | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CTGTGCTTCGTCCGCTCGGC.......................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CGCGGGACTCAGACCAGC....................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................AGAGTGTGGCCCGGGGC................................................................................................................................................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GAAGGGCGCCTGTGCT....................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| GCGGGCTCCGAGAAGAAAGAGTGTGGCCCGGGGCGGGCGCATTAGGAGGTGTCGAGGCCGTGGCCCGGAAGTGGTCGGGGCCGCTGCGGGCGGAAGGGCGCCTGTGCTTCGTCCGCTCGGCGGTGGCCCAGCCAGGCCCGCGGGACTCAGACCAGCGGGGAGCGCGACCTCCGCCCTTGGGGCTCTCCCGCTGGG .....................................................((((.......)))).(((((((((((((((((.(((((....)))))..)))))))).)))))))))..................................................... ..........................................................59.......................................................................132............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR040016(GSM532901) G645N. (cervix) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040034(GSM532919) G001N. (cervix) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR207110(GSM721072) Nuclear RNA. (cell line) | GSM532876(GSM532876) G547T. (cervix) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR139184(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | TAX577742(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR139183(SRX050632) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR040019(GSM532904) G701T. (cervix) | SRR139167(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189782 | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR139192(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029125(GSM416754) U2OS. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................ccggCAGCGGGGAGCGCGAC........................... | 20 | ccgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................tttgGTGGCCCGGAAGTGG......................................................................................................................... | 19 | tttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......ggaGAGAAGAAAGAGTGT........................................................................................................................................................................... | 18 | gga | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................ccgtCTGTGCTTCGTCCGC............................................................................... | 19 | ccgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............GAAAGAGTGTGGCCCGGGGCGG............................................................................................................................................................... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ............................................................TGGCCCGGAAGTGGT........................................................................................................................ | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |