| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (2) B-CELL | (3) BRAIN | (10) BREAST | (18) CELL-LINE | (4) CERVIX | (5) FIBROBLAST | (1) HELA | (11) LIVER | (2) OTHER | (1) RRP40.ip | (32) SKIN | (1) SPLEEN | (1) XRN.ip |
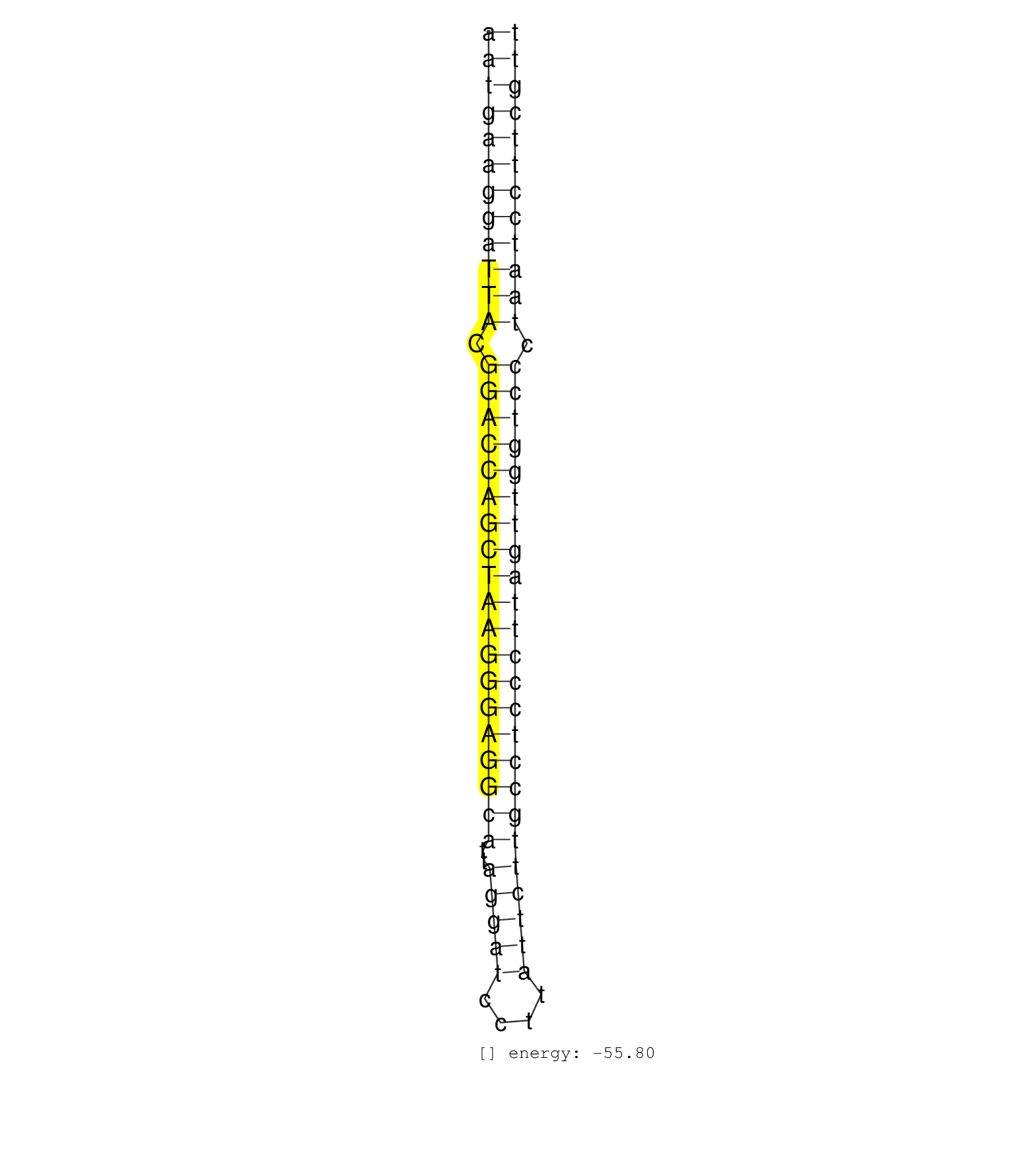
| TCTAAATTTGATGAAATGGCAAAGGTGGATAAAGTATGCTATGATTGGGAAATGAAGGATTACGGACCAGCTAAGGGAGGCATTAGGATCCTTATTCTTGCCTCCCTTAGTTGGTCCCTAATCCTTCGTTTGGATCCCACTGTCTGGATTCTTCCTGCTCTGTTCCAAATTCTGCCCTAA ..................................................((((((((((((.(((((((((((((((((((..(((((....)))))))))))))))))))))))).)))))))))))).................................................. ..................................................51.............................................................................130................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR189782 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | TAX577741(Rovira) total RNA. (breast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR139202(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040036(GSM532921) G243N. (cervix) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040025(GSM532910) G613T. (cervix) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | TAX577742(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR040011(GSM532896) G529T. (cervix) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191621(GSM715731) 16genomic small RNA (size selected RNA from t. (breast) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | TAX577739(Rovira) total RNA. (breast) | TAX577590(Rovira) total RNA. (breast) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR191582(GSM715692) 95genomic small RNA (size selected RNA from t. (breast) | TAX577738(Rovira) total RNA. (breast) | SRR038853(GSM458536) MELB. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................TTACGGACCAGCTAAGGGAGG.................................................................................................... | 21 | 2 | 10.50 | 10.50 | - | - | 5.00 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCATatc.............................................................................................. | 26 | ATC | 10.00 | 8.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAG..................................................................................................... | 20 | 2 | 8.50 | 8.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | 0.50 | - | - | - | - | - | - | - | 1.00 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGA...................................................................................................... | 19 | 2 | 8.50 | 8.50 | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCAT................................................................................................. | 23 | 1 | 8.00 | 8.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCAT................................................................................................. | 24 | 1 | 6.00 | 6.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGatc................................................................................................. | 24 | ATC | 6.00 | 10.50 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCA.................................................................................................. | 23 | 2 | 4.50 | 4.50 | - | - | - | 1.00 | 0.50 | - | - | - | - | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGC................................................................................................... | 22 | 2 | 4.00 | 4.00 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ACGGACCAGCTAAGGGAGGCAT................................................................................................. | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGa................................................................................................... | 22 | A | 3.50 | 10.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - |
| ............................................................TACGGACCAGCTAAGGGAG..................................................................................................... | 19 | 2 | 3.00 | 3.00 | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ......................................................AAGGATTACGGACCAGCTAAGGGAGGCAag................................................................................................ | 30 | AG | 3.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCATatc.............................................................................................. | 27 | ATC | 3.00 | 6.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCATaatc............................................................................................. | 28 | AATC | 3.00 | 6.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................GGAAATGAAGGATTACatg.................................................................................................................. | 19 | ATG | 3.00 | 0.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................ATGAAGGATTACGGACCAGCTAAGGGAGGCA.................................................................................................. | 31 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................ATGAAGGATTACGGACCAGCTAAGGGAGGCAa................................................................................................. | 32 | A | 2.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCATT................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ACGGACCAGCTAAGGGAGGCATT................................................................................................ | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCt.................................................................................................. | 22 | T | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCATTAaa............................................................................................. | 27 | AA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................AGGATTACGGACCAGCTAAGGGAGGCA.................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGtc................................................................................................... | 22 | TC | 2.00 | 8.50 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ACGGACCAGCTAAGGGAGGCATatc.............................................................................................. | 25 | ATC | 2.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCATa................................................................................................ | 25 | A | 2.00 | 6.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGa.................................................................................................... | 21 | A | 1.50 | 8.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCA.................................................................................................. | 22 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCATTt............................................................................................... | 25 | T | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CGGACCAGCTAAGGGAGGCA.................................................................................................. | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ACGGACCAGCTAAGGGAGGCAat................................................................................................ | 23 | AT | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGGGAAATGAAGGATggc..................................................................................................................... | 18 | GGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGGGAAATGAAGGATTttg.................................................................................................................... | 19 | TTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCg.................................................................................................. | 22 | G | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGt................................................................................................... | 22 | T | 1.00 | 10.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGaa................................................................................................... | 22 | AA | 1.00 | 8.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCATT................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CGGACCAGCTAAGGGAGGCAT................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGGCAat................................................................................................ | 24 | AT | 1.00 | 1.50 | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAagct.................................................................................................. | 22 | AGCT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AAGGATTACGGACCAGCTAAGGGAGGCA.................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGaa.................................................................................................. | 23 | AA | 1.00 | 10.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| .............................................................ACGGACCAGCTAAGGGAGGCATa................................................................................................ | 23 | A | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCCACTGTCTGGATTtctg.......................... | 19 | TCTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................AAATGAAGGATTACGGACCAGCT............................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................ATTACGGACCAGCTAAGGGAGGCA.................................................................................................. | 24 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGt.................................................................................................... | 20 | T | 0.50 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGaaa.................................................................................................. | 23 | AAA | 0.50 | 8.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAa..................................................................................................... | 20 | A | 0.50 | 8.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGtt.................................................................................................. | 23 | TT | 0.50 | 10.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CGGACCAGCTAAGGGAG..................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCAat................................................................................................ | 25 | AT | 0.50 | 4.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCAaa................................................................................................ | 25 | AA | 0.50 | 4.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAaa.................................................................................................... | 21 | AA | 0.50 | 8.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CGGACCAGCTAAGGGAGGCAaa................................................................................................ | 22 | AA | 0.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGgat................................................................................................. | 24 | GAT | 0.50 | 10.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................ACGGACCAGCTAAGGGAGGC................................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAGGCAg................................................................................................. | 24 | G | 0.50 | 4.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAt..................................................................................................... | 20 | T | 0.50 | 8.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGG....................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TACGGACCAGCTAAGGGAGG.................................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTACGGACCAGCTAAGGGAta.................................................................................................... | 21 | TA | 0.50 | 8.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTAAATTTGATGAAATGGCAAAGGTGGATAAAGTATGCTATGATTGGGAAATGAAGGATTACGGACCAGCTAAGGGAGGCATTAGGATCCTTATTCTTGCCTCCCTTAGTTGGTCCCTAATCCTTCGTTTGGATCCCACTGTCTGGATTCTTCCTGCTCTGTTCCAAATTCTGCCCTAA ..................................................((((((((((((.(((((((((((((((((((..(((((....)))))))))))))))))))))))).)))))))))))).................................................. ..................................................51.............................................................................130................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR189782 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | TAX577741(Rovira) total RNA. (breast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR139202(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040036(GSM532921) G243N. (cervix) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040025(GSM532910) G613T. (cervix) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | TAX577742(Rovira) total RNA. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR040011(GSM532896) G529T. (cervix) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191621(GSM715731) 16genomic small RNA (size selected RNA from t. (breast) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | TAX577739(Rovira) total RNA. (breast) | TAX577590(Rovira) total RNA. (breast) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR191582(GSM715692) 95genomic small RNA (size selected RNA from t. (breast) | TAX577738(Rovira) total RNA. (breast) | SRR038853(GSM458536) MELB. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................tcaTTCCTGCTCTGTTCC.............. | 18 | tca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................ccGATCCTTATTCTTGC............................................................................... | 17 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................gccCTGTTCCAAATTCTG...... | 18 | gcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |