| (2) B-CELL | (1) BRAIN | (10) BREAST | (1) CELL-LINE | (4) CERVIX | (1) FIBROBLAST | (8) HEART | (3) LIVER | (51) SKIN | (2) UTERUS |
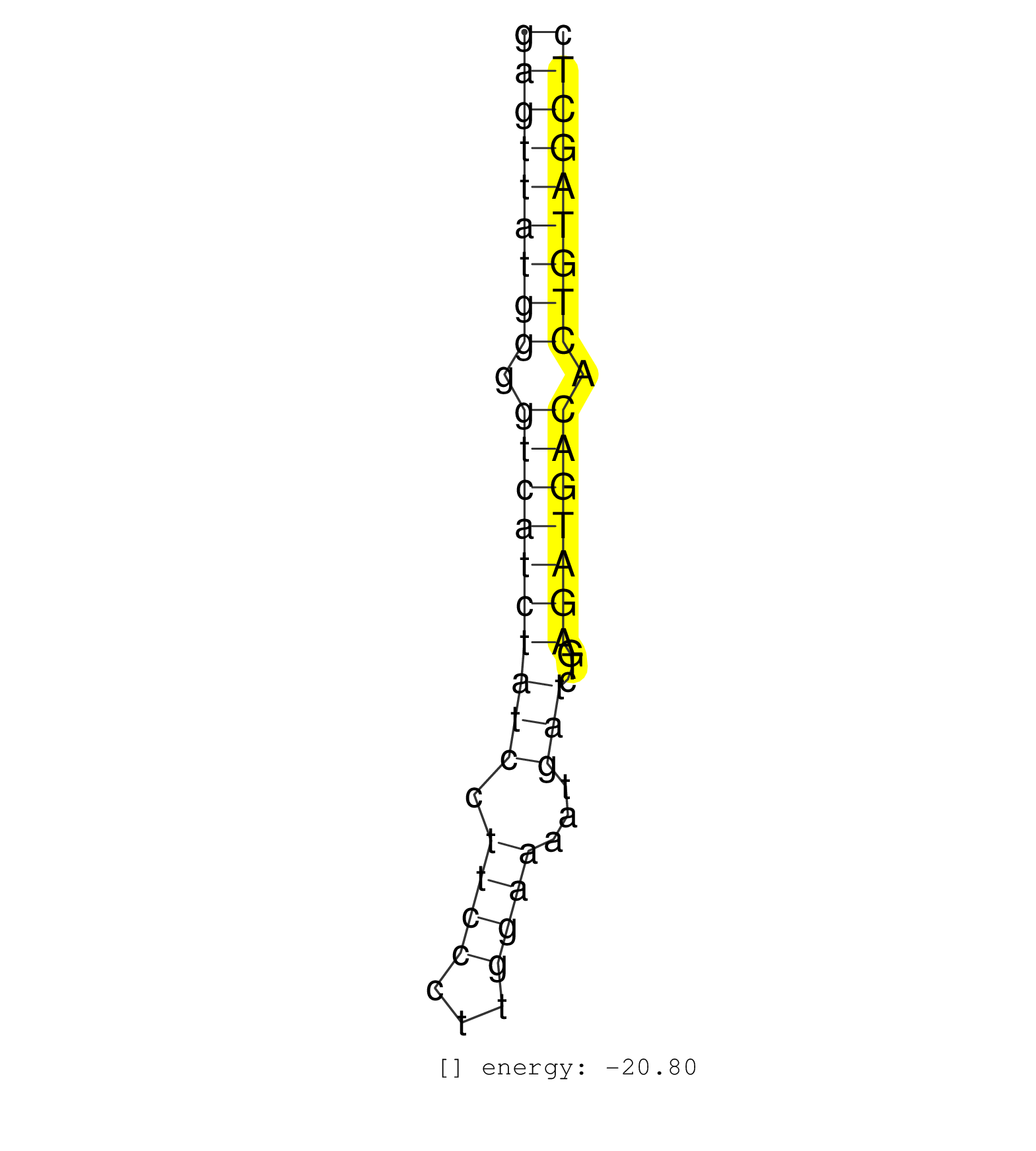
| GTTTTGATTCCTATCTTGCATGTGGAATCTCATTCTCTCTTAGAGATAATGAGTTATGGGGTCATCTATCCTTCCCTTGGAAAATGATCTGAGATGACACTGTAGCTCCCCATCCTCTGGGATCATTGATGTCTGTAAGATTTTATAGGAAGCAATTG ..................................................(((((((((.((((((((((.((((...))))...)))...))))))).))))))))).................................................. ..................................................51.......................................................108................................................ | Size | Perfect hit | Total Norm | Perfect Norm | TAX577580(Rovira) total RNA. (breast) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | TAX577742(Rovira) total RNA. (breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | TAX577588(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | TAX577741(Rovira) total RNA. (breast) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR040024(GSM532909) G613N. (cervix) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR040030(GSM532915) G013N. (cervix) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191592(GSM715702) 59genomic small RNA (size selected RNA from t. (breast) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040014(GSM532899) G623N. (cervix) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | TAX577453(Rovira) total RNA. (breast) | SRR040035(GSM532920) G001T. (cervix) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR038852(GSM458535) QF1160MB. (cell line) | TAX577744(Rovira) total RNA. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................TGAGATGACACTGTAGCTCt................................................. | 20 | T | 140.00 | 16.00 | - | 7.00 | 11.00 | 9.00 | 8.00 | 10.00 | - | 3.00 | 4.00 | 6.00 | 4.00 | 5.00 | 1.00 | 5.00 | 5.00 | - | 4.00 | 1.00 | 2.00 | 1.00 | 2.00 | 4.00 | 3.00 | 1.00 | 2.00 | - | 3.00 | 2.00 | 1.00 | - | 3.00 | 3.00 | - | 1.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | 2.00 | 1.00 | - | 1.00 | 1.00 | - | 2.00 | 1.00 | 1.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCT................................................... | 18 | 1 | 91.00 | 91.00 | 34.00 | 4.00 | 2.00 | 3.00 | 1.00 | - | 8.00 | 1.00 | - | - | 2.00 | 1.00 | 3.00 | - | - | 3.00 | - | 3.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 2.00 | - | 1.00 | 2.00 | - | 1.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTC.................................................. | 19 | 1 | 16.00 | 16.00 | - | 3.00 | - | - | 1.00 | - | 1.00 | 1.00 | 3.00 | - | - | - | 1.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGC.................................................... | 17 | 1 | 14.00 | 14.00 | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTCtt................................................ | 21 | TT | 6.00 | 16.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTCa................................................. | 20 | A | 5.00 | 16.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................TGAGATGACACTGTAGCTt.................................................. | 19 | T | 4.00 | 91.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCa................................................... | 18 | A | 3.00 | 14.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTa.................................................. | 19 | A | 3.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTg.................................................. | 19 | G | 2.00 | 91.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTCtc................................................ | 21 | TC | 2.00 | 16.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GCATGTGGAATCTCAaaa........................................................................................................................... | 18 | AAA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................CTTGGAAAATGATCTGAGATGACA........................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................AATGATCTGAGATGAagc.......................................................... | 18 | AGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................TGAGATGACACTGTAa..................................................... | 16 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTat................................................. | 20 | AT | 1.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTaa................................................. | 20 | AA | 1.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTCat................................................ | 21 | AT | 1.00 | 16.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CTGAGATGACACTGTAGCTCt................................................. | 21 | T | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GAGATGACACTGTAGCTCa................................................. | 19 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTCta................................................ | 21 | TA | 1.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTCC................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAG..................................................... | 16 | 4 | 1.00 | 1.00 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................AGAGATAATGAGTTATctg.................................................................................................. | 19 | CTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTCg................................................. | 20 | G | 1.00 | 16.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CTGAGATGACACTGTAGC.................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTgaa................................................ | 21 | GAA | 1.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGCTtt................................................. | 20 | TT | 1.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................TGAGATGACACTGTAGaaa.................................................. | 19 | AAA | 0.50 | 1.00 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| .........................................................................................TGAGATGACACTGTAGaaaa................................................. | 20 | AAAA | 0.25 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTTTTGATTCCTATCTTGCATGTGGAATCTCATTCTCTCTTAGAGATAATGAGTTATGGGGTCATCTATCCTTCCCTTGGAAAATGATCTGAGATGACACTGTAGCTCCCCATCCTCTGGGATCATTGATGTCTGTAAGATTTTATAGGAAGCAATTG ..................................................(((((((((.((((((((((.((((...))))...)))...))))))).))))))))).................................................. ..................................................51.......................................................108................................................ | Size | Perfect hit | Total Norm | Perfect Norm | TAX577580(Rovira) total RNA. (breast) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | TAX577742(Rovira) total RNA. (breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | TAX577588(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330873(SRX091711) tissue: skin psoriatic involveddisease state:. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | TAX577741(Rovira) total RNA. (breast) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR040024(GSM532909) G613N. (cervix) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR040030(GSM532915) G013N. (cervix) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191592(GSM715702) 59genomic small RNA (size selected RNA from t. (breast) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040014(GSM532899) G623N. (cervix) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | TAX577453(Rovira) total RNA. (breast) | SRR040035(GSM532920) G001T. (cervix) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR038852(GSM458535) QF1160MB. (cell line) | TAX577744(Rovira) total RNA. (breast) |
|---|