| (1) AGO1.ip OTHER.mut | (7) B-CELL | (1) BREAST | (17) CELL-LINE | (1) HELA | (1) KIDNEY | (9) LIVER | (1) OTHER | (4) SKIN | (3) UTERUS | (1) XRN.ip |
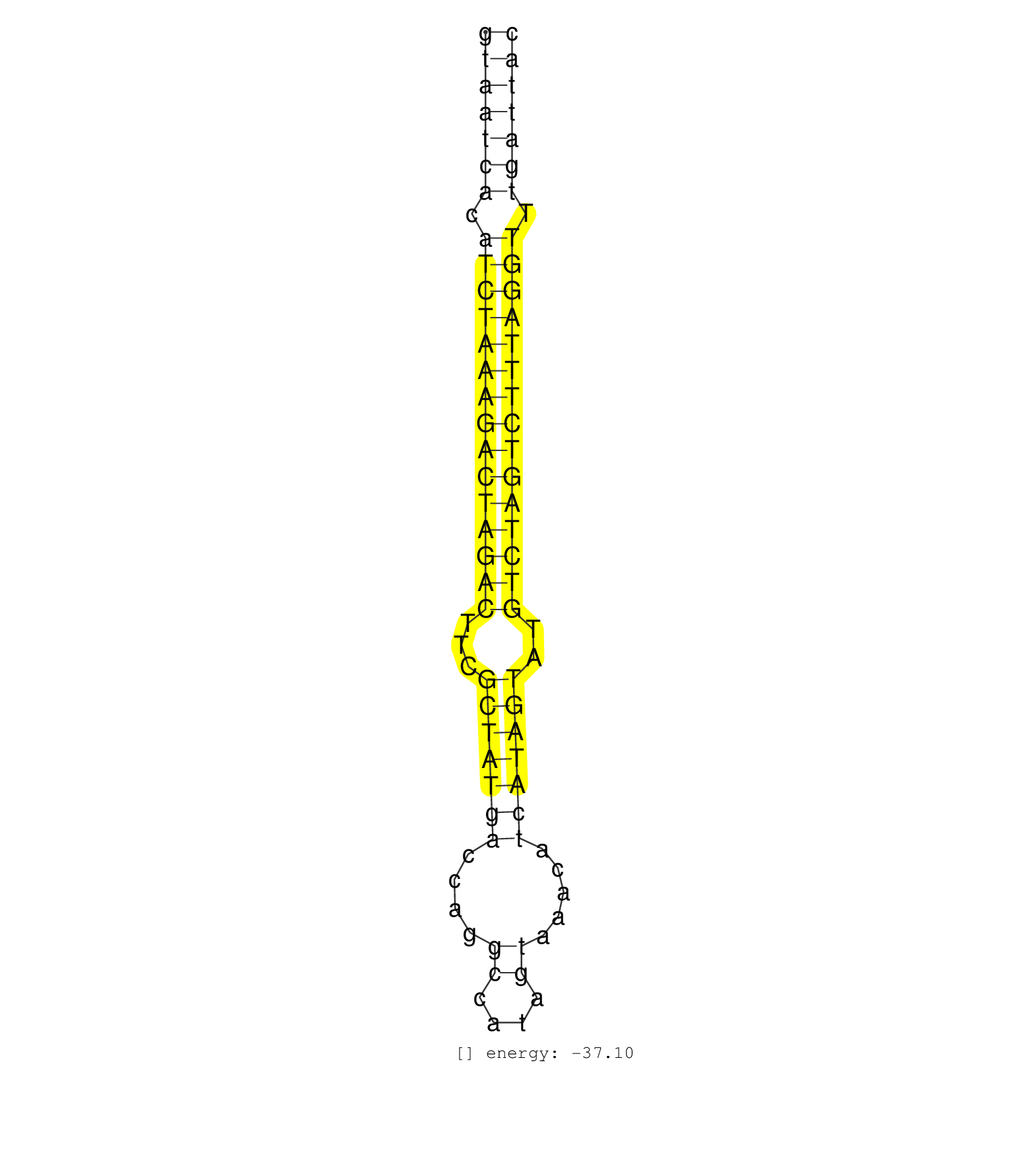
| TATCCATTTGGGTGCATAAGAGAAATGGGCTTGTAAGAGAAATGGGCTCTGTAATCACATCTAAAGACTAGACTTCGCTATGACCAGGCCATAGTAAACATCATAGTATGTCTAGTCTTTAGGTTTGATTACAGAACATTATAAGAGCATCCCTTATACTTGCATGTCATTTTAATGTCAGT ..................................................(((((((.(((((((((((((((...(((((((....((....)).....)))))))..))))))))))))))).))))))).................................................. ..................................................51...............................................................................132................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037936(GSM510474) 293cand1. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR015358(GSM380323) Naïve B Cell (Naive39). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | TAX577739(Rovira) total RNA. (breast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037935(GSM510473) 293cand3. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139176(SRX050650) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR038857(GSM458540) D20. (cell line) | SRR189786 | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR038856(GSM458539) D11. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................TCTAAAGACTAGACTTCGCTAT..................................................................................................... | 22 | 1 | 15.00 | 15.00 | 2.00 | - | 1.00 | 2.00 | - | 1.00 | - | 1.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGCTATG.................................................................................................... | 23 | 1 | 14.00 | 14.00 | - | 1.00 | - | - | - | 1.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGCTATGA................................................................................................... | 24 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGCTA...................................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................ATAGTATGTCTAGTCTTTAGGTT......................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAGTATGTCTAGTCTTTAGGT.......................................................... | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGCTATc.................................................................................................... | 23 | C | 2.00 | 15.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TAAAGACTAGACTTCGCTAT..................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGCTATGACCAGGCC............................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TTAGGTTTGATTACAcacc............................................. | 19 | CACC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGattt..................................................................................................... | 22 | ATTT | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TAAAGACTAGACTTCGCTATG.................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGCTATGAa.................................................................................................. | 25 | A | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAGTATGTCTAGTCTTTAGG........................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATAGTATGTCTAGTCTTcagg........................................................... | 21 | CAGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CTAAAGACTAGACTTCGCTATGA................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGCT....................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TCTGTAATCACATCTAAAGA................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTAAAGACTAGACTTCGCTATGt................................................................................................... | 24 | T | 1.00 | 14.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGCATCCCTTATACTTaa................... | 18 | AA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................TCTGTAATCACATCTAAAGACT................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................AAATGGGCTTGTAAG................................................................................................................................................. | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| ..........................GGGCTTGTAAGAGAA............................................................................................................................................. | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| TATCCATTTGGGTGCATAAGAGAAATGGGCTTGTAAGAGAAATGGGCTCTGTAATCACATCTAAAGACTAGACTTCGCTATGACCAGGCCATAGTAAACATCATAGTATGTCTAGTCTTTAGGTTTGATTACAGAACATTATAAGAGCATCCCTTATACTTGCATGTCATTTTAATGTCAGT ..................................................(((((((.(((((((((((((((...(((((((....((....)).....)))))))..))))))))))))))).))))))).................................................. ..................................................51...............................................................................132................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037936(GSM510474) 293cand1. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR015358(GSM380323) Naïve B Cell (Naive39). (B cell) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | TAX577739(Rovira) total RNA. (breast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037935(GSM510473) 293cand3. (cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139176(SRX050650) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR029128(GSM416757) H520. (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR038857(GSM458540) D20. (cell line) | SRR189786 | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR038856(GSM458539) D11. (cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................tcaCCATAGTAAACATCATAG............................................................................ | 21 | tca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |