| (1) B-CELL | (3) BRAIN | (4) BREAST | (10) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (1) HEART | (1) HELA | (15) LIVER | (4) OTHER | (4) OVARY | (1) PLACENTA | (6) SKIN | (2) TESTES | (1) UTERUS |
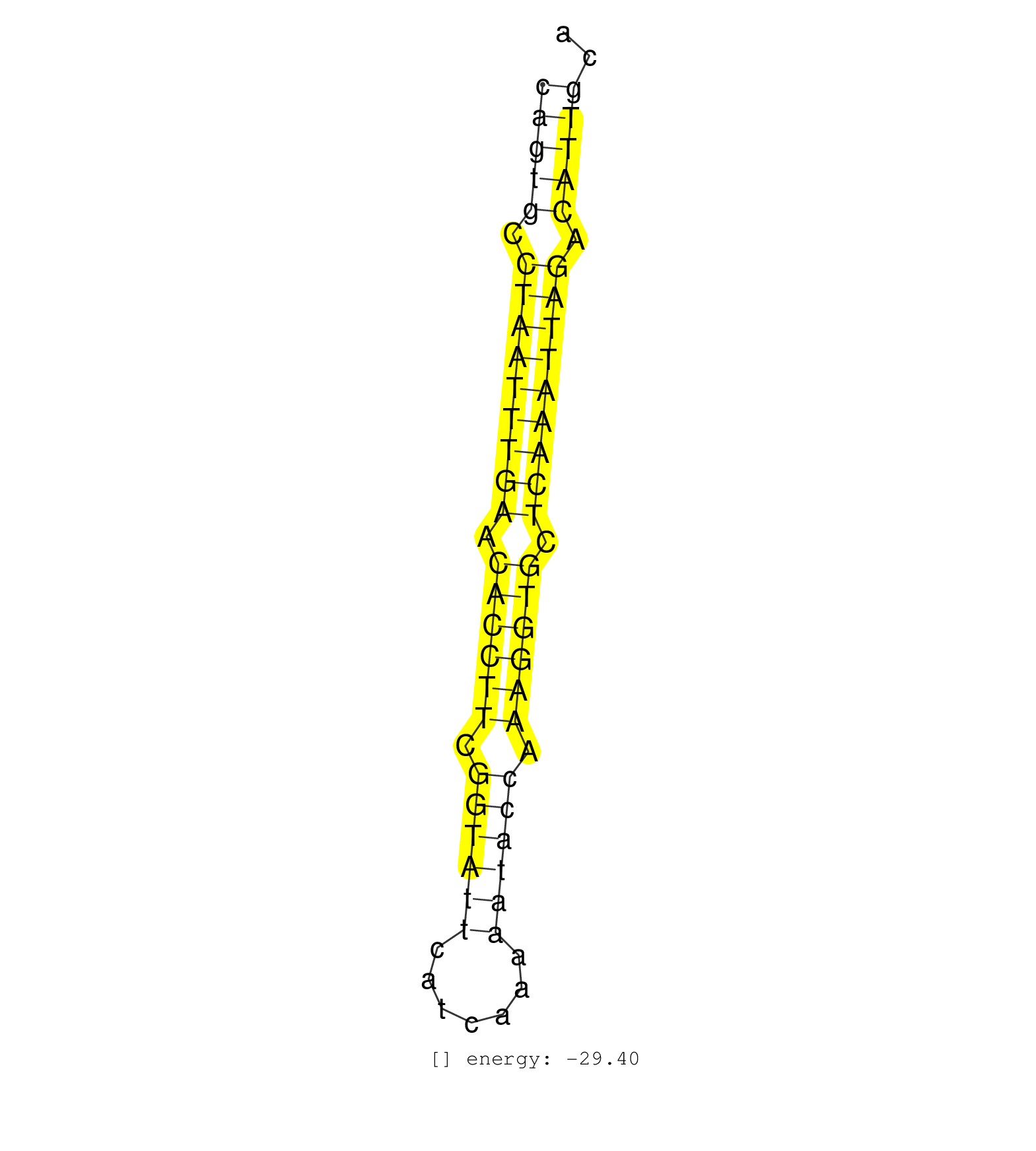
| AAAATCTCAGCTCCACTTCTGCAAGTTTATCTTCCCTGCTTGCATAAAATTGCAGTGCCTAATTTGAACACCTTCGGTATTCATCAAAAATACCAAAGGTGCTCAAATTAGACATTGCATTTTTTGCGCATGTCAAGTTGAAATCTATTCATGGATCTTTAGCTCCTTA ..................................................(((((.(((((((((.((((((.((((((.......)))))).)))))).))))))))).))))).................................................... ....................................................53................................................................119................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR139179(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139177(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR189782 | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR139178(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | TAX577741(Rovira) total RNA. (breast) | SRR139180(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029124(GSM416753) HeLa. (hela) | SRR037936(GSM510474) 293cand1. (cell line) | SRR139191(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR037938(GSM510476) 293Red. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR189784 | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191457(GSM715567) 27genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139198(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR189785 | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR139209(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139192(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR038859(GSM458542) MM386. (cell line) | SRR139211(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR040012(GSM532897) G648N. (cervix) | SRR189786 | TAX577589(Rovira) total RNA. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR139193(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | TAX577738(Rovira) total RNA. (breast) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................AAAGGTGCTCAAATTAGACATT..................................................... | 22 | 1 | 59.00 | 59.00 | 21.00 | 15.00 | 13.00 | 3.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCGGTAa......................................................................................... | 23 | A | 42.00 | 23.00 | - | - | - | - | 5.00 | 3.00 | 3.00 | 3.00 | 2.00 | 3.00 | 3.00 | - | - | - | 2.00 | - | 2.00 | 3.00 | 1.00 | 3.00 | 1.00 | - | 1.00 | - | - | 2.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCGGTA.......................................................................................... | 22 | 1 | 23.00 | 23.00 | - | - | - | - | - | - | 1.00 | - | 3.00 | - | - | - | 3.00 | 3.00 | - | - | 1.00 | - | 2.00 | - | 2.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - |
| .........................................................CCTAATTTGAACACCTTCGGTAT......................................................................................... | 23 | 1 | 11.00 | 11.00 | - | - | - | - | - | 1.00 | 2.00 | 1.00 | - | 1.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCG............................................................................................. | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................AAGGTGCTCAAATTAGACATT..................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCGGT........................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - |
| .........................................................CCTAATTTGAACACCTTCGGTAaa........................................................................................ | 24 | AA | 2.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................AAAGGTGCTCAAATTAGACA....................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AAAGGTGCTCAAATTAGACAT...................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCGG............................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCaacg.......................................................................................... | 22 | AACG | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCGata.......................................................................................... | 22 | ATA | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CTAATTTGAACACCTTCGGTAa......................................................................................... | 22 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCtgta.......................................................................................... | 22 | TGTA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CCAAAGGTGCTCAAATTAGACt....................................................... | 22 | T | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CCAAAGGTGCTCAAATTAGACA....................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................AAAGGTGCTCAAATTAGACAct..................................................... | 22 | CT | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCGcgc.......................................................................................... | 22 | CGC | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCGacg.......................................................................................... | 22 | ACG | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CCAAAGGTGCTCAAATTAGAC........................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CAGCTCCACTTCTGCAAGgttt............................................................................................................................................ | 22 | GTTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CCTAATTTGAACACCTTCGtgcc......................................................................................... | 23 | TGCC | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CCAAAGGTGCTCAAATTAGA......................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ACCAAAGGTGCTCAAATTAG.......................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CATCAAAAATACCAAcg....................................................................... | 17 | CG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAAATCTCAGCTCCACTTCTGCAAGTTTATCTTCCCTGCTTGCATAAAATTGCAGTGCCTAATTTGAACACCTTCGGTATTCATCAAAAATACCAAAGGTGCTCAAATTAGACATTGCATTTTTTGCGCATGTCAAGTTGAAATCTATTCATGGATCTTTAGCTCCTTA ..................................................(((((.(((((((((.((((((.((((((.......)))))).)))))).))))))))).))))).................................................... ....................................................53................................................................119................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR139179(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139177(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR189782 | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR139178(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | TAX577741(Rovira) total RNA. (breast) | SRR139180(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029124(GSM416753) HeLa. (hela) | SRR037936(GSM510474) 293cand1. (cell line) | SRR139191(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR037938(GSM510476) 293Red. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR139194(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR189784 | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191457(GSM715567) 27genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139198(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR189785 | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR139209(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139192(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR038859(GSM458542) MM386. (cell line) | SRR139211(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR040012(GSM532897) G648N. (cervix) | SRR189786 | TAX577589(Rovira) total RNA. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR139193(SRX050634) FLASHPage purified small RNA (~15-40nt) from . (ovary) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | TAX577738(Rovira) total RNA. (breast) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................agcgGTCAAGTTGAAATCT....................... | 19 | agcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .gcggCTCAGCTCCACTTCT..................................................................................................................................................... | 19 | gcgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....taaAGCTCCACTTCTGCA.................................................................................................................................................. | 18 | taa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................caaGTCAAGTTGAAATCT....................... | 18 | caa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................gGCCTAATTTGAACAC.................................................................................................. | 16 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |