| (1) AGO2.ip | (6) BREAST | (8) CELL-LINE | (1) HEART | (12) LIVER | (1) LUNG | (1) OTHER | (1) SKIN | (1) TESTES |
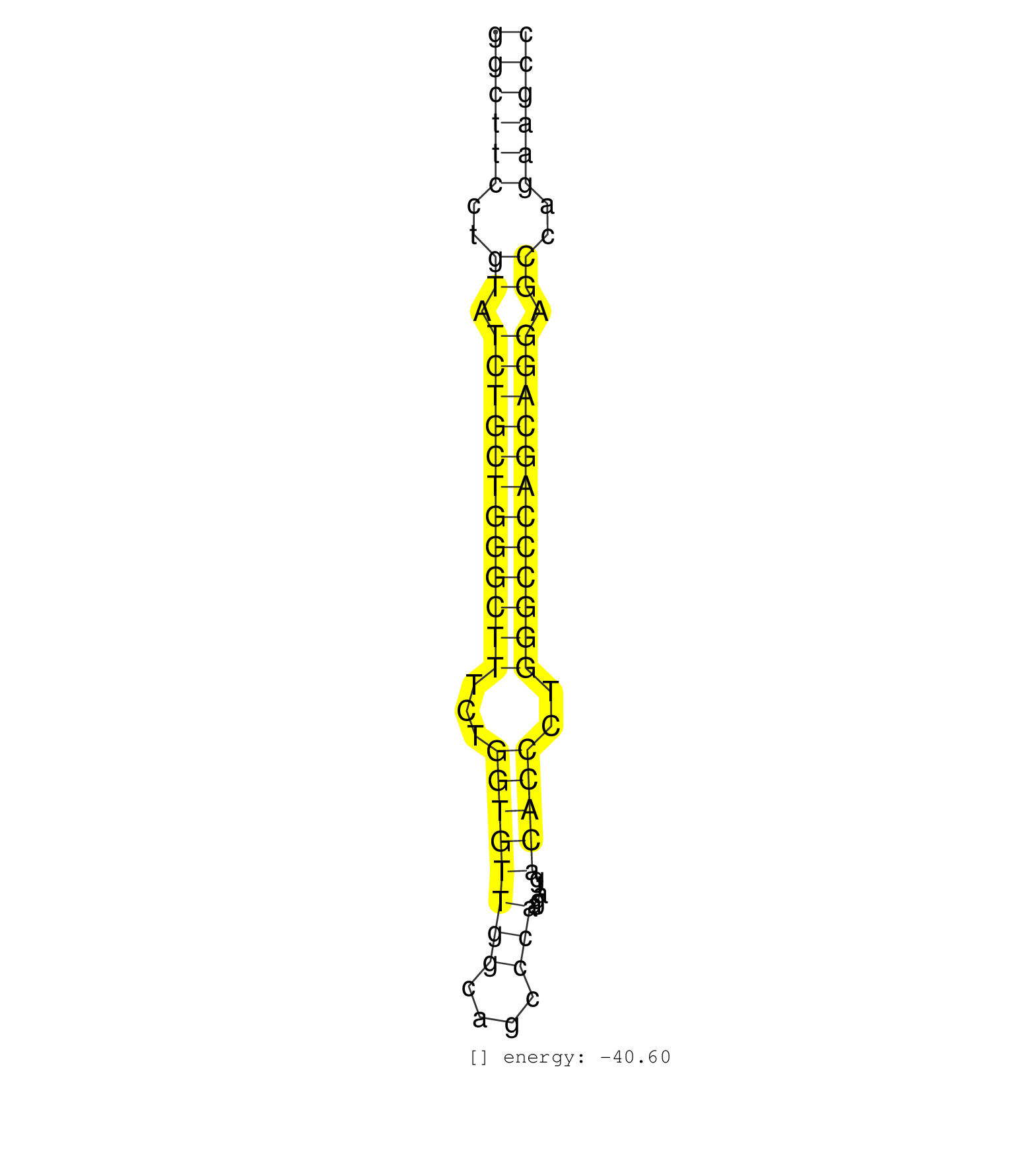
| GCTGCCACCGCTGTCCCTCCTGGAGAAGAACCGTTTCCGACAGGGCTTGGGGCTTCCTGTATCTGCTGGGCTTTCTGGTGTTGGCAGCCCAAGATGACACCCTGGGCCCAGCAGGAGCCAGAAGCCCTAGACGCTCCCTGACTTCTCGAAGGCCTACCCCGGCTGGTCACTGTGGC ..................................................((((((..((.((((((((((((...((((((((....))).....)))))..)))))))))))).))..)))))).................................................. ..................................................51.........................................................................126................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR029129(GSM416758) SW480. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | TAX577739(Rovira) total RNA. (breast) | SRR029131(GSM416760) MCF7. (cell line) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR139179(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR139188(SRX050640) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | TAX577743(Rovira) total RNA. (breast) | SRR189782 | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................TATCTGCTGGGCTTTCTGGTGTT.............................................................................................. | 23 | 1 | 50.00 | 50.00 | 8.00 | 3.00 | 2.00 | 8.00 | 6.00 | 6.00 | 2.00 | 3.00 | 4.00 | - | 2.00 | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGT............................................................................................... | 22 | 1 | 13.00 | 13.00 | 3.00 | 1.00 | 2.00 | - | - | - | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTG................................................................................................ | 21 | 1 | 8.00 | 8.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGa............................................................................................... | 22 | A | 4.00 | 8.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGTTt............................................................................................. | 24 | T | 4.00 | 50.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGTTa............................................................................................. | 24 | A | 4.00 | 50.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGT................................................................................................. | 20 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGG.................................................................................................. | 19 | 2 | 2.50 | 2.50 | 1.00 | 1.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTa................................................................................................ | 21 | A | 2.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGTa.............................................................................................. | 23 | A | 2.00 | 13.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGTTag............................................................................................ | 25 | AG | 2.00 | 50.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CAGCAGGAGCCAGAAcagg................................................. | 19 | CAGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGTTGt............................................................................................ | 25 | T | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGg............................................................................................... | 22 | G | 1.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TCCCTCCTGGAGAAGAccc................................................................................................................................................ | 19 | CCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCgagc................................................................................................. | 20 | GAGC | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGTTatt........................................................................................... | 26 | ATT | 1.00 | 50.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................ACACCCTGGGCCCAGCAGGAat.......................................................... | 22 | AT | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGc............................................................................................... | 22 | C | 1.00 | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGTTG............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CTGCCACCGCTGTCCCcc............................................................................................................................................................. | 18 | CC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGTGTag............................................................................................. | 24 | AG | 1.00 | 13.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TCCCTGACTTCTCGAcaac....................... | 19 | CAAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGcg................................................................................................ | 21 | CG | 0.50 | 2.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGagtt.............................................................................................. | 23 | AGTT | 0.50 | 2.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTGGgg................................................................................................ | 21 | GG | 0.50 | 2.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATCTGCTGGGCTTTCTG................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .................................................................................................CACCCTGGGCCCAGCAGGAGC.......................................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| GCTGCCACCGCTGTCCCTCCTGGAGAAGAACCGTTTCCGACAGGGCTTGGGGCTTCCTGTATCTGCTGGGCTTTCTGGTGTTGGCAGCCCAAGATGACACCCTGGGCCCAGCAGGAGCCAGAAGCCCTAGACGCTCCCTGACTTCTCGAAGGCCTACCCCGGCTGGTCACTGTGGC ..................................................((((((..((.((((((((((((...((((((((....))).....)))))..)))))))))))).))..)))))).................................................. ..................................................51.........................................................................126................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR029129(GSM416758) SW480. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | TAX577739(Rovira) total RNA. (breast) | SRR029131(GSM416760) MCF7. (cell line) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | TAX577579(Rovira) total RNA. (breast) | SRR139179(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR139188(SRX050640) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | TAX577743(Rovira) total RNA. (breast) | SRR189782 | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................ttgGAAGCCCTAGACGCT......................................... | 18 | ttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................atAGAAGCCCTAGACGCT......................................... | 18 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................ATCTGCTGGGCTTTCTG................................................................................................... | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |