| (1) AGO2.ip | (7) B-CELL | (3) BRAIN | (2) BREAST | (14) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (2) HEART | (1) HELA | (4) LIVER | (1) OTHER | (1) PLACENTA | (3) SKIN | (2) UTERUS |
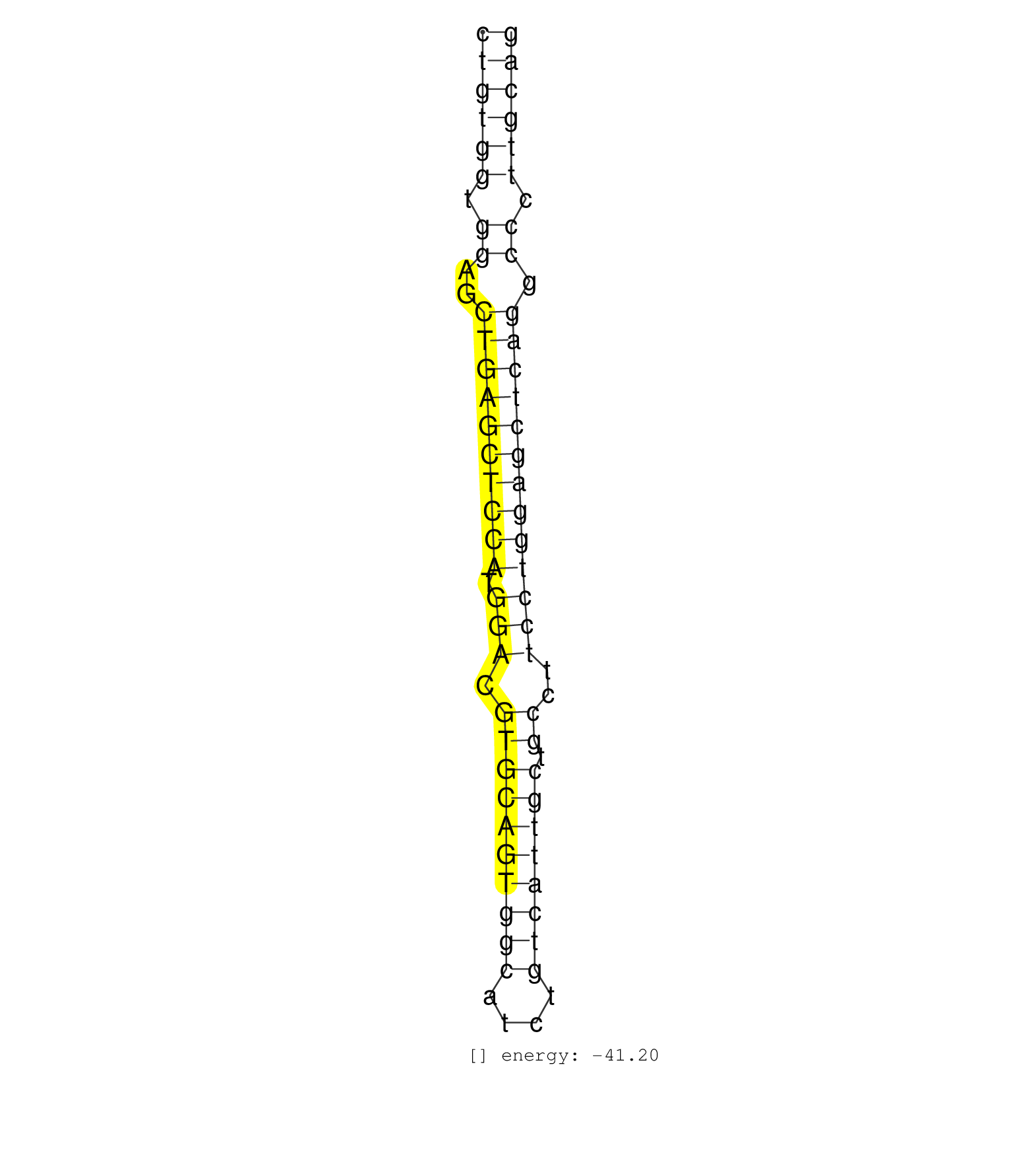
| TTCCTCCTGGCCAGATCACAAGCTCTAGGGCGGTGTCTCTGGTTTGCCTTCTGTGGTGGAGCTGAGCTCCATGGACGTGCAGTGGCATCTGTCATTGCTGCCTTCCTGGAGCTCAGGCCCTTGCAGAGGCAGTCTGAGGCATGGCAAGCCACAAGCGCACAGCCTGTCCCCACCAA ..................................................((((((.((..((((((((((.(((.((((((((((....)))))))).))..))))))))))))).)).)))))).................................................. ..................................................51.........................................................................126................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR040018(GSM532903) G701N. (cervix) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | TAX577742(Rovira) total RNA. (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR343335 | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189786 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR038863(GSM458546) MM603. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................AGCTGAGCTCCATGGACGTGCAGT............................................................................................. | 24 | 1 | 27.00 | 27.00 | - | 4.00 | 3.00 | 2.00 | 2.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - |
| ...........................................................AGCTGAGCTCCATGGACGTGCAG.............................................................................................. | 23 | 1 | 9.00 | 9.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - |
| ...........................................................AGCTGAGCTCCATGGACGTGCA............................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................AGCTGAGCTCCATGGACGTGCAGatc........................................................................................... | 26 | ATC | 2.00 | 9.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................GCGGTGTCTCTGGTTgag................................................................................................................................. | 18 | GAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TCCATGGACGTGCAGT............................................................................................. | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGCTGAGCTCCATGGACGTGCAGTat........................................................................................... | 26 | AT | 1.00 | 27.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGCTGAGCTCCATGGACGTGa................................................................................................ | 21 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................ATCTGTCATTGCTGCtaca....................................................................... | 19 | TACA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCTAGGGCGGTGTCTCTGG...................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................ATGGACGTGCAGTGGC.......................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................AGCTGAGCTCCATGGACGTGC................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGGAGCTCAGGCCCTTcccg.................................................. | 20 | CCCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGCTGAGCTCCATGGACGTGCAGc............................................................................................. | 24 | C | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ATGGCAAGCCACAAGCGCACAGCCTGTCCCC..... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................GAGCTGAGCTCCATGGACGTGCAG.............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGCTGAGCTCCATGGACGTGgag.............................................................................................. | 23 | GAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................CCTTCTGTGGTGGAGC.................................................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| TTCCTCCTGGCCAGATCACAAGCTCTAGGGCGGTGTCTCTGGTTTGCCTTCTGTGGTGGAGCTGAGCTCCATGGACGTGCAGTGGCATCTGTCATTGCTGCCTTCCTGGAGCTCAGGCCCTTGCAGAGGCAGTCTGAGGCATGGCAAGCCACAAGCGCACAGCCTGTCCCCACCAA ..................................................((((((.((..((((((((((.(((.((((((((((....)))))))).))..))))))))))))).)).)))))).................................................. ..................................................51.........................................................................126................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR040018(GSM532903) G701N. (cervix) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | TAX577742(Rovira) total RNA. (breast) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR343335 | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189786 | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR038863(GSM458546) MM603. (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................cTCATTGCTGCCTTCCTG.................................................................... | 18 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................tCCTTGCAGAGGCAGTCT......................................... | 18 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................agGGTGTCTCTGGTTTGC................................................................................................................................. | 18 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |