| (2) AGO2.ip | (2) B-CELL | (11) BREAST | (26) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (3) HEART | (1) HELA | (1) KIDNEY | (3) LIVER | (1) OTHER | (17) SKIN | (1) TESTES | (3) THYMUS | (1) UTERUS | (1) XRN.ip |
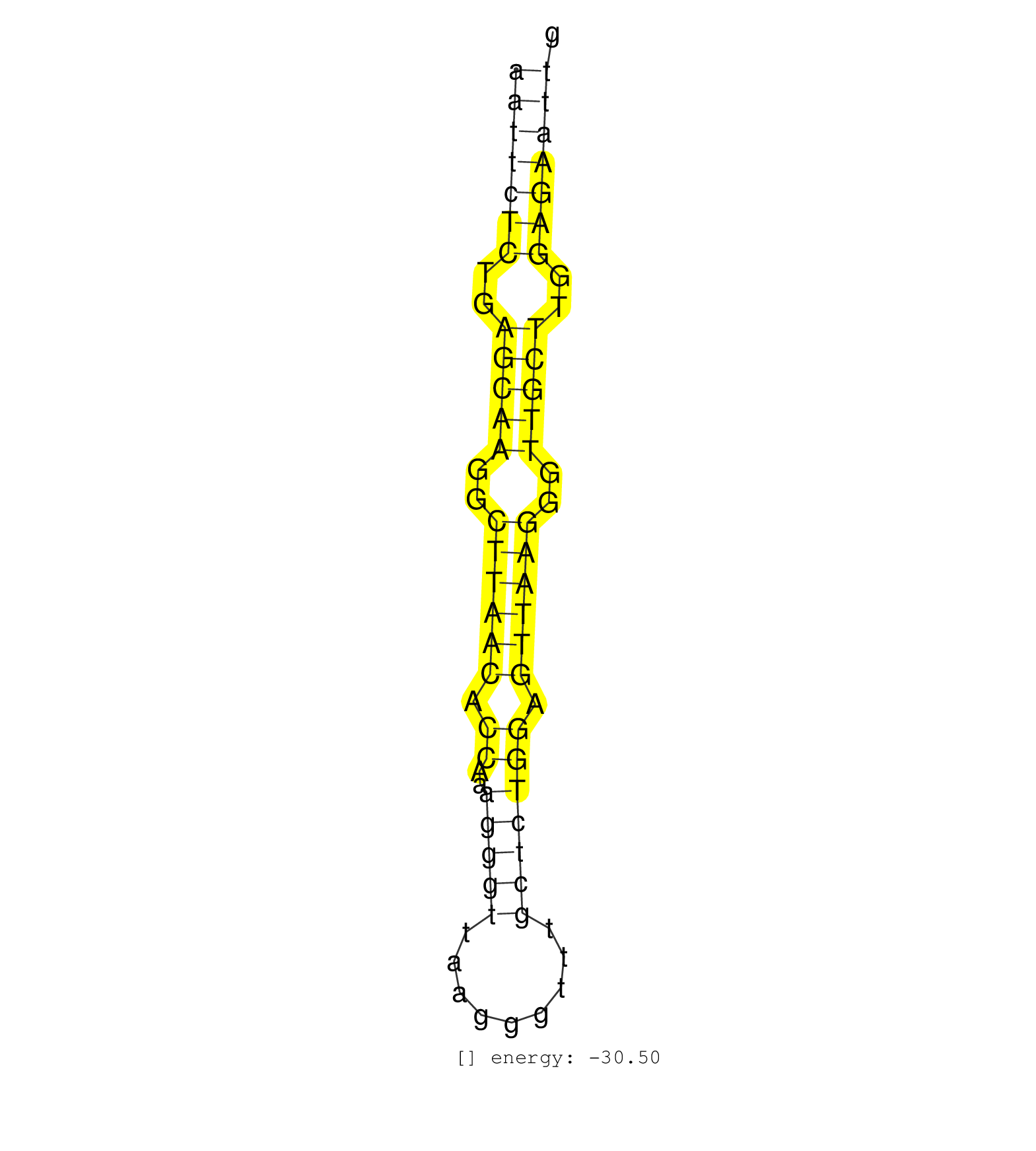
| CTCTGTCCTTCTCCCAAGCCCCCAGATGTCTCGTGGCCACCCAGTCTCCTTTGTCCAATTCTCTGAGCAAGGCTTAACACCAAAGGGTTAAGGGTTTGCTCTGGAGTTAAGGGTTGCTTGGAGAATTGGAGAAGGGGCACCTGGTAGGTCATTCTTTTCTTCCCTGATTTCTTCCCCCACCTC ..................................................(((((((..(((((..((((((.((..(((((.........))))))).))))))..)))))..)))))))................................................... ........................................................57.....................................................................128..................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR139215(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR029128(GSM416757) H520. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189782 | SRR139214(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR038857(GSM458540) D20. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR139165(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191543(GSM715653) 124genomic small RNA (size selected RNA from . (breast) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR040028(GSM532913) G026N. (cervix) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | GSM532874(GSM532874) G699T. (cervix) | GSM416733(GSM416733) HEK293. (cell line) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR038855(GSM458538) D10. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR037931(GSM510469) 293GFP. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR038858(GSM458541) MEL202. (cell line) | SRR139177(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR038859(GSM458542) MM386. (cell line) | SRR139211(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR189786 | SRR191560(GSM715670) 77genomic small RNA (size selected RNA from t. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................TGGAGTTAAGGGTTGCTTGGAGA........................................................... | 23 | 1 | 15.00 | 15.00 | 2.00 | - | - | 4.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................TCTGAGCAAGGCTTAACACCAgt................................................................................................... | 23 | GT | 9.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................TCTGAGCAAGGCTTAACACCA..................................................................................................... | 21 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGAGCAAGGCTTAACACCAgag.................................................................................................. | 22 | GAG | 8.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GGAGTTAAGGGTTGCTTGGAGA........................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GAGTTAAGGGTTGCTTGGAGA........................................................... | 21 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGGAGTTAAGGGTTGCTTGGAG............................................................ | 22 | 1 | 4.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................TCTGAGCAAGGCTTAACACC...................................................................................................... | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................GAGTTAAGGGTTGCTTGGAGAA.......................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TCTGAGCAAGGCTTAACACCAga................................................................................................... | 23 | GA | 3.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................CTGGAGTTAAGGGTTGCTTGGA............................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGGAGTTAAGGGTTGCTTGGAa............................................................ | 22 | A | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGGAGTTAAGGGTTGCTTGGAGAA.......................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGAGCAAGGCTTAACACCAgaga................................................................................................. | 23 | GAGA | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GAGTTAAGGGTTGCTTGGAG............................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................TCTGAGCAAGGCTTAACACCAg.................................................................................................... | 22 | G | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTCTGAGCAAGGCTTAACACC...................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGAGCAAGGCTTAACACCAga................................................................................................... | 21 | GA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGCTTGGAGAATTGGAGA................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTCTGAGCAAGGCTTAACA........................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TCTGAGCAAGGCTTAACACCAA.................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................TGGAGTTAAGGGTTGCTTGGtga........................................................... | 23 | TGA | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GAGTTAAGGGTTGCTTGGAtat.......................................................... | 22 | TAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GGAGTTAAGGGTTGCTTGGAG............................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GGGTTAAGGGTTTGCTCTGGAGTTAAGGGT..................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TTGCTTGGAGAATTGGAGAA.................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................ATTCTTTTCTTCCCTGcagc............. | 20 | CAGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGGAGTTAAGGGTTGCTTGGAGAt.......................................................... | 24 | T | 1.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCATTCTTTTCTTCCtcg................. | 18 | TCG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................TTCTTTTCTTCCCTGgacg............. | 19 | GACG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCTCTGAGCAAGGCTTAACAC....................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GAGTTAAGGGTTGCTTGGAGAt.......................................................... | 22 | T | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GAGTTAAGGGTTGCTTGGAGAAa......................................................... | 23 | A | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TCTGAGCAAGGCTTAACAC....................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CTGGAGTTAAGGGTTGCTTGG.............................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGGAGTTAAGGGTTGCTTGGAGg........................................................... | 23 | G | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GAGTTAAGGGTTGCTTGG.............................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TTGCTTGGAGAATTGGA..................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGAGCAAGGCTTAACACCAAAG.................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TCTGAGCAAGGCTTAACACCAAt................................................................................................... | 23 | T | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGGAGTTAAGGGTTGCTT................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| .....................................................................................................................TTGGAGAATTGGAGAAG................................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTCTGTCCTTCTCCCAAGCCCCCAGATGTCTCGTGGCCACCCAGTCTCCTTTGTCCAATTCTCTGAGCAAGGCTTAACACCAAAGGGTTAAGGGTTTGCTCTGGAGTTAAGGGTTGCTTGGAGAATTGGAGAAGGGGCACCTGGTAGGTCATTCTTTTCTTCCCTGATTTCTTCCCCCACCTC ..................................................(((((((..(((((..((((((.((..(((((.........))))))).))))))..)))))..)))))))................................................... ........................................................57.....................................................................128..................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR029130(GSM416759) DLD2. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR139215(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR029128(GSM416757) H520. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR189782 | SRR139214(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR038857(GSM458540) D20. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR139165(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191543(GSM715653) 124genomic small RNA (size selected RNA from . (breast) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR040028(GSM532913) G026N. (cervix) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | GSM532874(GSM532874) G699T. (cervix) | GSM416733(GSM416733) HEK293. (cell line) | SRR330909(SRX091747) tissue: normal skindisease state: normal. (skin) | SRR038855(GSM458538) D10. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR139171(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR037931(GSM510469) 293GFP. (cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR038858(GSM458541) MEL202. (cell line) | SRR139177(SRX050638) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR038859(GSM458542) MM386. (cell line) | SRR139211(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR189786 | SRR191560(GSM715670) 77genomic small RNA (size selected RNA from t. (breast) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................ccatCTTTGTCCAATTCTCT....................................................................................................................... | 20 | ccat | 134.00 | 0.00 | 93.00 | 11.00 | 12.00 | - | - | 5.00 | 4.00 | 6.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................catCTTTGTCCAATTCTCT....................................................................................................................... | 19 | cat | 21.00 | 0.00 | 4.00 | 12.00 | 1.00 | - | - | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................ccatCTTTGTCCAATTCTC........................................................................................................................ | 19 | ccat | 5.00 | 0.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................atCTTTGTCCAATTCTCT....................................................................................................................... | 18 | at | 4.00 | 0.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................tcatCTTTGTCCAATTCTCT....................................................................................................................... | 20 | tcat | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................gccTTGTCCAATTCTCTG...................................................................................................................... | 18 | gcc | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................cagaTGTCCAATTCTCTGAG.................................................................................................................... | 20 | caga | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................catCTTTGTCCAATTCTC........................................................................................................................ | 18 | cat | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................ccagCTTTGTCCAATTCTCT....................................................................................................................... | 20 | ccag | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................ccagCTTTGTCCAATTCTC........................................................................................................................ | 19 | ccag | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................ccaCCTTTGTCCAATTCTCT....................................................................................................................... | 20 | cca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................acatCTTTGTCCAATTCTCT....................................................................................................................... | 20 | acat | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................CTTTGTCCAATTCTCTGAGCAAGG............................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................attTTTTCTTCCCTGATTTC............ | 20 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................cgttTTGGAGAAGGGGCACCT......................................... | 21 | cgtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGTCCAATTCTCTGAGCAAGG............................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |