| (2) BRAIN | (20) BREAST | (6) CELL-LINE | (4) CERVIX | (1) FIBROBLAST | (4) HEART | (1) OTHER | (46) SKIN | (1) TESTES | (1) THYMUS |
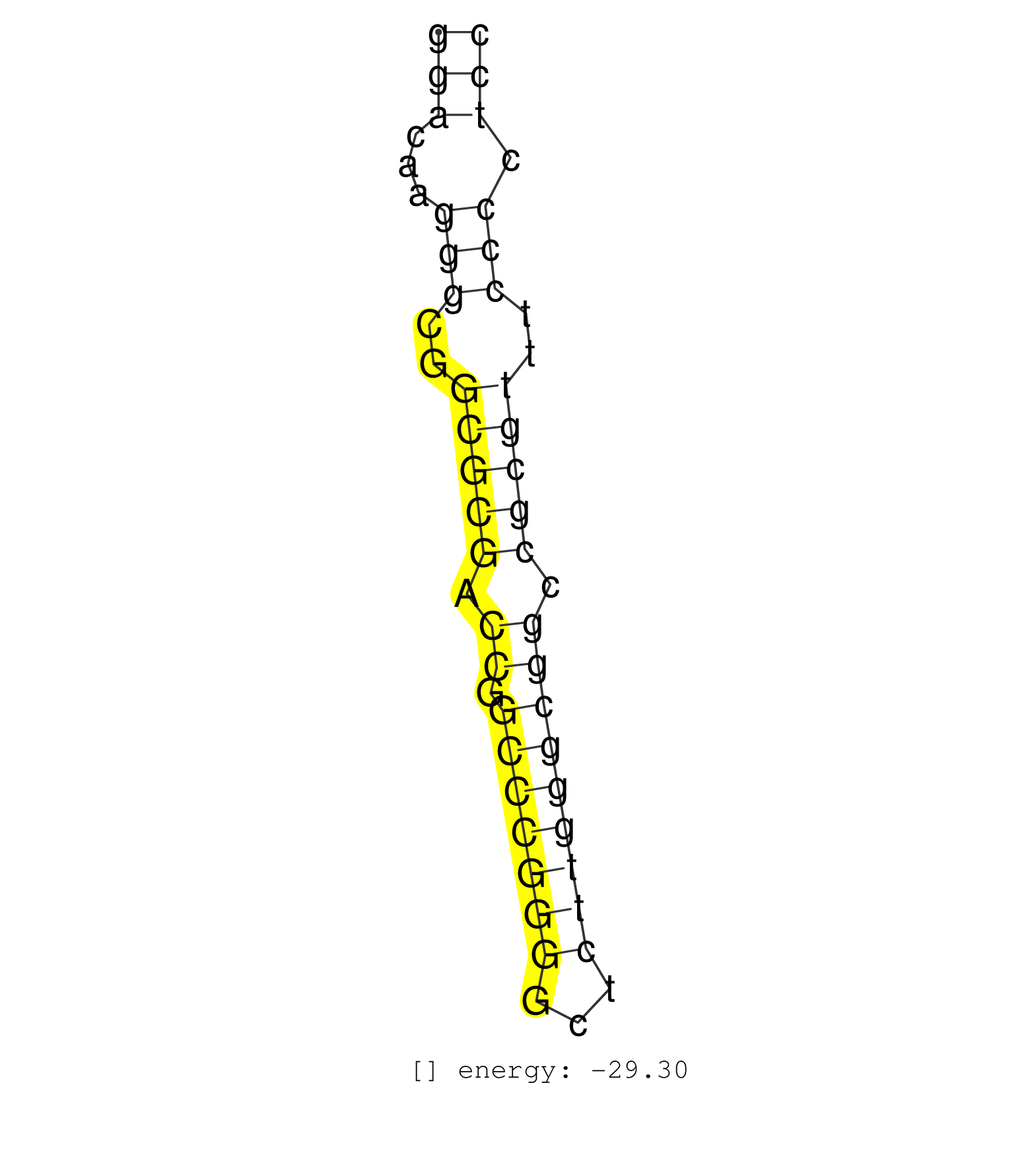
| GCCGCGCGCCCCGCCATCAGCTGCGCGCACAGAGGGAGAGTTCACTCCGGGGACAAGGGCGGCGCGACCGGCCCGGGGCTCTTGGGCGGCCGCGTTTCCCCTCCCACCGGCTGCAGCCCCCTTCTCCCGCGGCACTTTCCGGCGTCTTTTGTGG ..................................................(((...(((..(((((.((.(((((((...))))))))).)))))..))).))).................................................. ..................................................51...................................................104................................................ | Size | Perfect hit | Total Norm | Perfect Norm | TAX577589(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189785 | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | TAX577590(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577740(Rovira) total RNA. (breast) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191554(GSM715664) 99genomic small RNA (size selected RNA from t. (breast) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577741(Rovira) total RNA. (breast) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR040018(GSM532903) G701N. (cervix) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | GSM532877(GSM532877) G691N. (cervix) | SRR040030(GSM532915) G013N. (cervix) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR191587(GSM715697) 50genomic small RNA (size selected RNA from t. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR139212(SRX050646) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037943(GSM510481) 293DcrTN. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR191631(GSM715741) 75genomic small RNA (size selected RNA from t. (breast) | SRR191403(GSM715513) 44genomic small RNA (size selected RNA from t. (breast) | TAX577739(Rovira) total RNA. (breast) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029126(GSM416755) 143B. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | TAX577453(Rovira) total RNA. (breast) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | TAX577745(Rovira) total RNA. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR444041(SRX128889) Sample 2cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................CGGCGCGACCGGCCCGGGG............................................................................ | 19 | 1 | 17.00 | 17.00 | 3.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCGGG............................................................................. | 18 | 1 | 15.00 | 15.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCGGG............................................................................. | 17 | 1 | 10.00 | 10.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCGG.............................................................................. | 16 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCGGGG............................................................................ | 18 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCGG.............................................................................. | 17 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCGGGGC........................................................................... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCG............................................................................... | 16 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................CGCGACCGGCCCGGGG............................................................................ | 16 | 2 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | - |
| ...........................................................CGGCGCGACCGGCCCccgg............................................................................ | 19 | CCGG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCcggg............................................................................ | 19 | CGGG | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCGGGGCaa......................................................................... | 21 | AA | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCcga............................................................................. | 18 | CGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCGGcg............................................................................ | 18 | CG | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCGGcggg.......................................................................... | 21 | CGGG | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCcccc............................................................................ | 19 | CCCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GCGCGACCGGCCCGGG............................................................................. | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GCGCGACCGGCCCGGGG............................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCGGcgga.......................................................................... | 21 | CGGA | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................GCCCGGGGCTCTTGGaaag................................................................. | 19 | AAAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CGCGACCGGCCCGGG............................................................................. | 15 | 5 | 0.60 | 0.60 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| GCCGCGCGCCCCGCCATCAGCTGCGCGCACAGAGGGAGAGTTCACTCCGGGGACAAGGGCGGCGCGACCGGCCCGGGGCTCTTGGGCGGCCGCGTTTCCCCTCCCACCGGCTGCAGCCCCCTTCTCCCGCGGCACTTTCCGGCGTCTTTTGTGG ..................................................(((...(((..(((((.((.(((((((...))))))))).)))))..))).))).................................................. ..................................................51...................................................104................................................ | Size | Perfect hit | Total Norm | Perfect Norm | TAX577589(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189785 | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | TAX577590(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577742(Rovira) total RNA. (breast) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577740(Rovira) total RNA. (breast) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR191554(GSM715664) 99genomic small RNA (size selected RNA from t. (breast) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577741(Rovira) total RNA. (breast) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR040018(GSM532903) G701N. (cervix) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | GSM532877(GSM532877) G691N. (cervix) | SRR040030(GSM532915) G013N. (cervix) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR191552(GSM715662) 42genomic small RNA (size selected RNA from t. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR191587(GSM715697) 50genomic small RNA (size selected RNA from t. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR139216(SRX050633) FLASHPage purified small RNA (~15-40nt) from . (thymus) | SRR139212(SRX050646) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037943(GSM510481) 293DcrTN. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR191631(GSM715741) 75genomic small RNA (size selected RNA from t. (breast) | SRR191403(GSM715513) 44genomic small RNA (size selected RNA from t. (breast) | TAX577739(Rovira) total RNA. (breast) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029126(GSM416755) 143B. (cell line) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | TAX577453(Rovira) total RNA. (breast) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | TAX577745(Rovira) total RNA. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR444041(SRX128889) Sample 2cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................CGGCGCGACCGGCCCGGGG............................................................................ | 19 | 1 | 23.00 | 23.00 | - | - | - | 1.00 | - | 3.00 | - | - | - | 1.00 | 3.00 | - | - | - | - | 2.00 | - | 3.00 | 1.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................GCGCGACCGGCCCGGGG............................................................................ | 17 | 1 | 17.00 | 17.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCGGGG............................................................................ | 18 | 1 | 13.00 | 13.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................aCCCGGGGCTCTTGGG.................................................................... | 16 | a | 7.00 | 0.00 | - | - | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ccgaCGGCGCGACCGGCCC................................................................................ | 19 | ccga | 6.00 | 0.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................aCGGCGCGACCGGCCCG............................................................................... | 17 | a | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ccgaCGGCGCGACCGGCCCGG.............................................................................. | 21 | ccga | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ccgaCGGCGCGACCGGCCCG............................................................................... | 20 | ccga | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCG............................................................................... | 16 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................gaCGGCGCGACCGGCCCG............................................................................... | 18 | ga | 2.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GCCCCCTTCTCCCGCGG...................... | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................gaCGGCGCGACCGGCCCGGG............................................................................. | 20 | ga | 2.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................aCGGCGCGACCGGCCCGGGG............................................................................ | 20 | a | 2.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GCGCGACCGGCCCGGGGC........................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GCGCGACCGGCCCGGG............................................................................. | 16 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................tacCCCCCTTCTCCCGCG....................... | 18 | tac | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCG............................................................................... | 15 | 2 | 1.50 | 1.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ...........................................................ctGCGCGACCGGCCCGGGG............................................................................ | 19 | ct | 1.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................cccaGCGGCGCGACCGGCCCG............................................................................... | 21 | ccca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCGGG............................................................................. | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................aCGGCGCGACCGGCCCGG.............................................................................. | 18 | a | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................cgaCGGCGCGACCGGCCCGGGG............................................................................ | 22 | cga | 1.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................GGCGCGACCGGCCCGGGGC........................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................cccGGCGGCGCGACCGGCCCG............................................................................... | 21 | ccc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................cccGGCGGCGCGACCGGC.................................................................................. | 18 | ccc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................acgaCGGCGCGACCGGCCC................................................................................ | 19 | acga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ccgaCGGCGCGACCGGCCCGGG............................................................................. | 22 | ccga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................aacCCCCCTTCTCCCGCG....................... | 18 | aac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................gcggAGCCCCCTTCTCCCG......................... | 19 | gcgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................gcaGCGGCGCGACCGGCC................................................................................. | 18 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GCCCCCTTCTCCCGCGGCACTTTCCGGCGT......... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................gaggGGGCGGCCGCGTTTCC....................................................... | 20 | gagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ccgaCGGCGCGACCGGCCCGGGG............................................................................ | 23 | ccga | 1.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................gaCGGCGCGACCGGCCCGGGG............................................................................ | 21 | ga | 1.00 | 23.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................cGGGCGGCGCGACCGGCCC................................................................................ | 19 | c | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................gaCGGCGCGACCGGCCCGG.............................................................................. | 19 | ga | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................cccGGCGGCGCGACCGGCCCGGGG............................................................................ | 24 | ccc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................ccGGGCGGCGCGACCGGCC................................................................................. | 19 | cc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ccaaCGGCGCGACCGGCCCGG.............................................................................. | 21 | ccaa | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................acaGCGCGACCGGCCCGGG............................................................................. | 19 | aca | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................aggGGGCGGCCGCGTTTCC....................................................... | 19 | agg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................CGGCGCGACCGGCCCGG.............................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CGCGACCGGCCCGGGG............................................................................ | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGGCGGCCGCGTTTC........................................................ | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ..............................................................CGCGACCGGCCCGGG............................................................................. | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................GCCCGGGGCTCTTGGG.................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |