| (2) B-CELL | (1) BREAST | (2) CELL-LINE | (3) CERVIX | (1) OTHER | (2) SKIN | (1) XRN.ip |
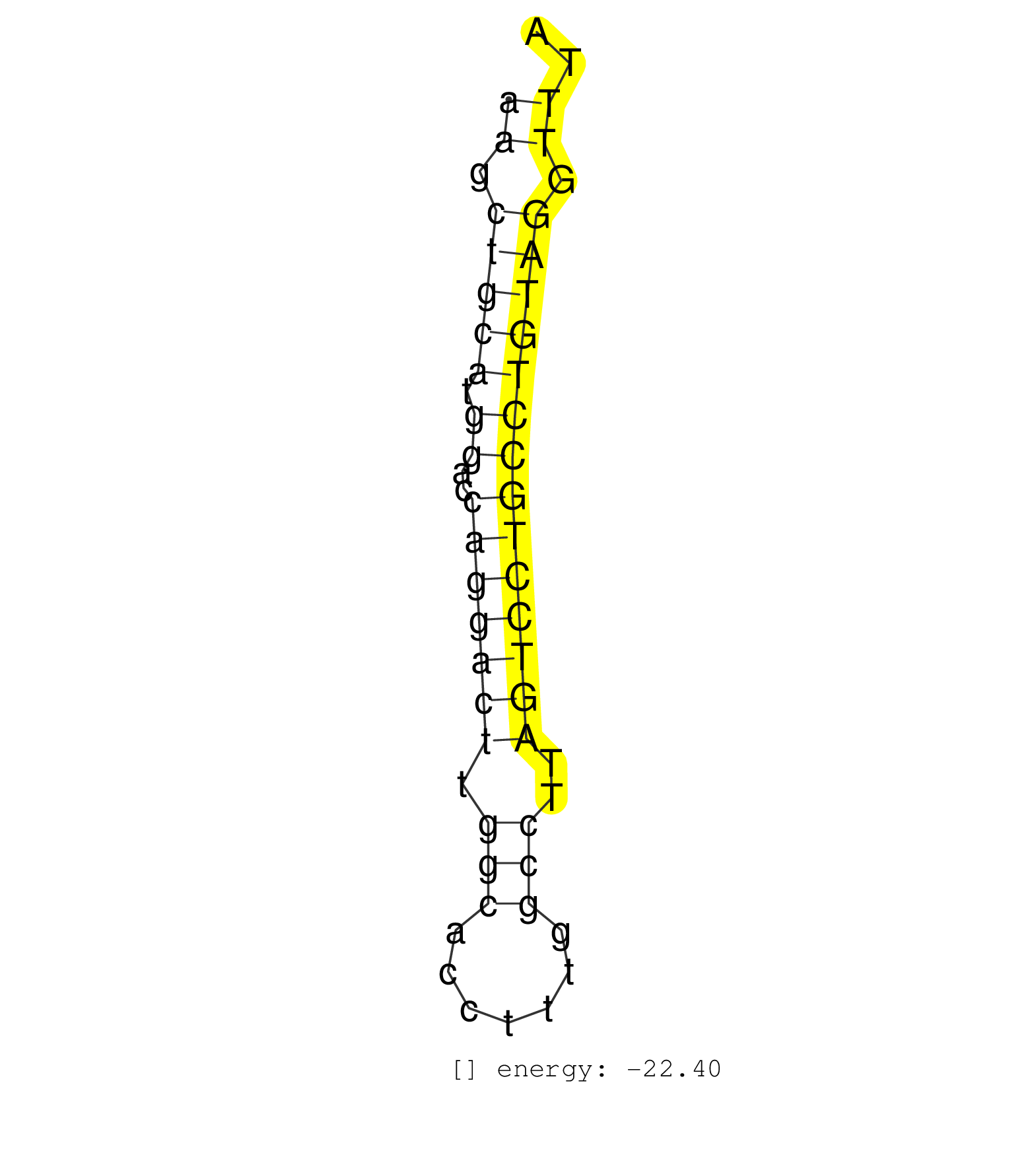
| GTCCACTCCTGGAACTCCTCAGGCTCTAGTGGAGAATTAGGGAGGCTGAGAAGCTGCATGGACCAGGACTTGGCACCTTTGGCCTTAGTCCTGCCTGTAGGTTTACCAGCCTTAGAGGGAAGTCACCTTTCTGTCTCTGTACCTCAGCCTGCACT ..................................................((.(((((.((..(((((((.(((.......)))..)))))))))))))).)).................................................... ..................................................51....................................................105................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040010(GSM532895) G529N. (cervix) | SRR189785 | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR040038(GSM532923) G531N. (cervix) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR040035(GSM532920) G001T. (cervix) | TAX577743(Rovira) total RNA. (breast) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................TTAGTCCTGCCTGTAGGTTTA.................................................. | 21 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................GAGAATTAGGGAGGCTGAGgt....................................................................................................... | 21 | GT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................AGGGAGGCTGAGAAGCacca................................................................................................. | 20 | ACCA | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........CTGGAACTCCTCAGGCTCTAGaa............................................................................................................................ | 23 | AA | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................ACCAGCCTTAGAGGGggat................................ | 19 | GGAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................CACCTTTGGCCTTAGTCCTGC............................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................GTCTCTGTACCTCAGCagg.... | 19 | AGG | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................ATTAGGGAGGCTGAGgtgc..................................................................................................... | 19 | GTGC | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....ACTCCTGGAACTCCTCttct................................................................................................................................... | 20 | TTCT | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................AGGCTGAGAAGCTGCA................................................................................................. | 16 | 6 | 0.17 | 0.17 | - | 0.17 | - | - | - | - | - | - | - | - | - |
| ...........................................................GGACCAGGACTTGGC................................................................................. | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | 0.11 |
| GTCCACTCCTGGAACTCCTCAGGCTCTAGTGGAGAATTAGGGAGGCTGAGAAGCTGCATGGACCAGGACTTGGCACCTTTGGCCTTAGTCCTGCCTGTAGGTTTACCAGCCTTAGAGGGAAGTCACCTTTCTGTCTCTGTACCTCAGCCTGCACT ..................................................((.(((((.((..(((((((.(((.......)))..)))))))))))))).)).................................................... ..................................................51....................................................105................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040010(GSM532895) G529N. (cervix) | SRR189785 | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR040038(GSM532923) G531N. (cervix) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR040035(GSM532920) G001T. (cervix) | TAX577743(Rovira) total RNA. (breast) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................tggaGTTTACCAGCCTTAGA....................................... | 20 | tgga | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |