| (3) B-CELL | (1) BRAIN | (2) BREAST | (1) CELL-LINE | (1) CERVIX | (3) HEART | (1) HELA | (1) LIVER | (1) OTHER | (5) SKIN |
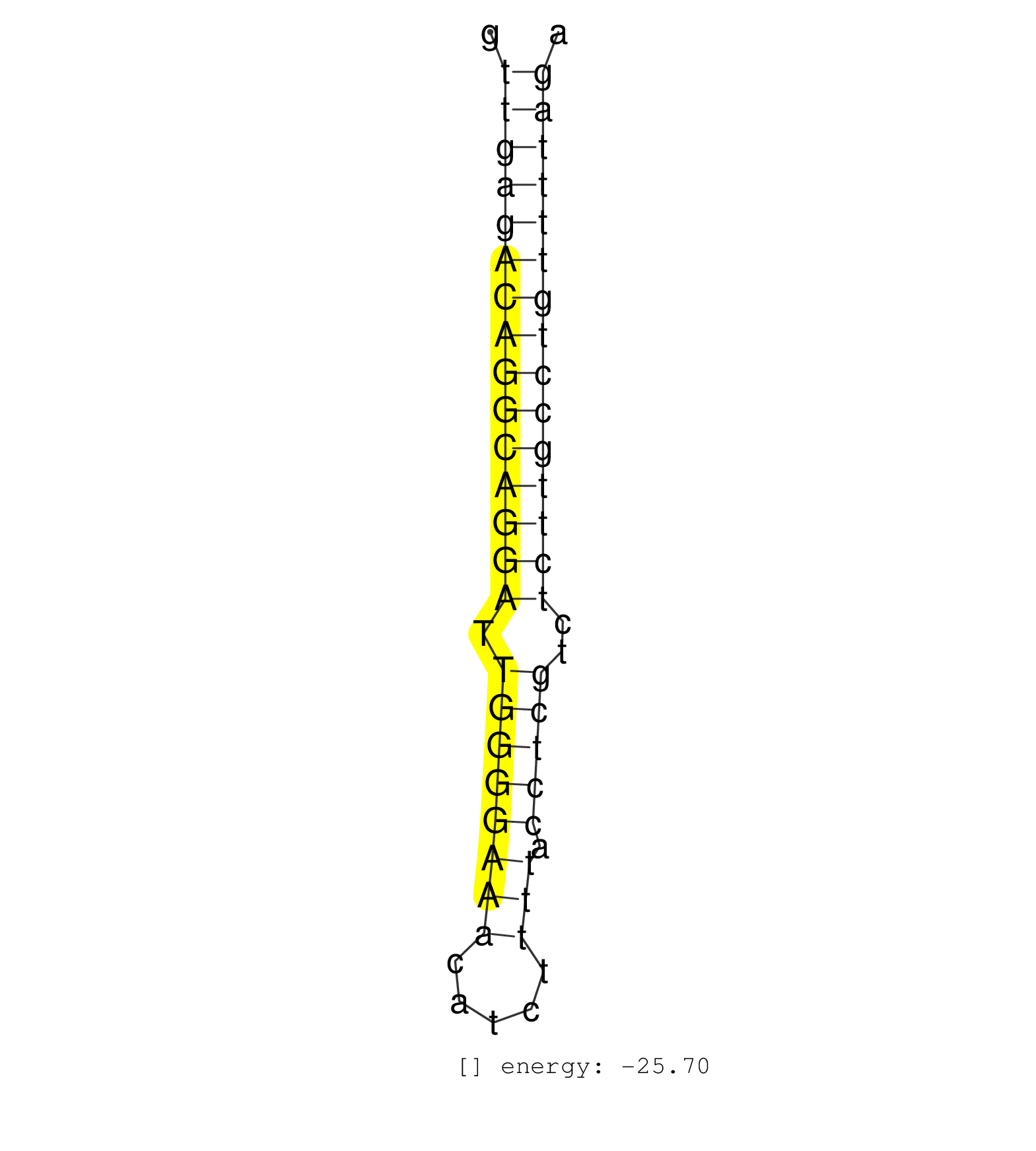
| AGTTGCAAAGAGAGAACTGAGGATTTGGGCAGGACAAGAGGAACTGACAGGTTGAGACAGGCAGGATTGGGGAAACATCTTTTACCTCGTCTCTTGCCTGTTTTAGAAGGTGCATCCACCACTTGGTATAAGAGAACATTATGGGCATTCTGTTTTG ...................................................(((((((((((((((.((((((((.....))).)))))..)))))))))))))))................................................... ..................................................51......................................................107................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033728(GSM497073) MALT (MALT413). (B cell) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR040010(GSM532895) G529N. (cervix) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR189787 | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | TAX577579(Rovira) total RNA. (breast) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................ACAGGCAGGATTGGGGAA................................................................................... | 18 | 2 | 11.50 | 11.50 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 0.50 | - | - |
| .............................CAGGACAAGAGGAACTctg............................................................................................................. | 19 | CTG | 2.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................ATCCACCACTTGGTAcca.......................... | 18 | CCA | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................GATTTGGGCAGGACAAGAG..................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGCAAAGAGAGAACTctg........................................................................................................................................ | 18 | CTG | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......AAGAGAGAACTGAGGATTTGGGC............................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AAGAGAACATTATGGGCA.......... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GAACATTATGGGCATTCTG..... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AGACAGGCAGGATTGtaga.................................................................................... | 19 | TAGA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................GAACATTATGGGCATTa....... | 17 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................ACAGGCAGGATTGGGGAAAa................................................................................. | 20 | A | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................GGCAGGACAAGAGGActc................................................................................................................ | 18 | CTC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTTGAGACAGGCAGG............................................................................................ | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - |
| ........................................................................................................................................CATTATGGGCATTCT...... | 15 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |
| AGTTGCAAAGAGAGAACTGAGGATTTGGGCAGGACAAGAGGAACTGACAGGTTGAGACAGGCAGGATTGGGGAAACATCTTTTACCTCGTCTCTTGCCTGTTTTAGAAGGTGCATCCACCACTTGGTATAAGAGAACATTATGGGCATTCTGTTTTG ...................................................(((((((((((((((.((((((((.....))).)))))..)))))))))))))))................................................... ..................................................51......................................................107................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033728(GSM497073) MALT (MALT413). (B cell) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR040010(GSM532895) G529N. (cervix) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR189787 | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | TAX577579(Rovira) total RNA. (breast) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577589(Rovira) total RNA. (breast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................ggtACAAGAGGAACTGACA............................................................................................................ | 19 | ggt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................tctcCTTGCCTGTTTTAGAA................................................. | 20 | tctc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |