| (22) B-CELL | (1) BRAIN | (7) BREAST | (10) CELL-LINE | (1) CERVIX | (1) FIBROBLAST | (1) HEART | (2) LIVER | (7) SKIN | (1) TESTES |
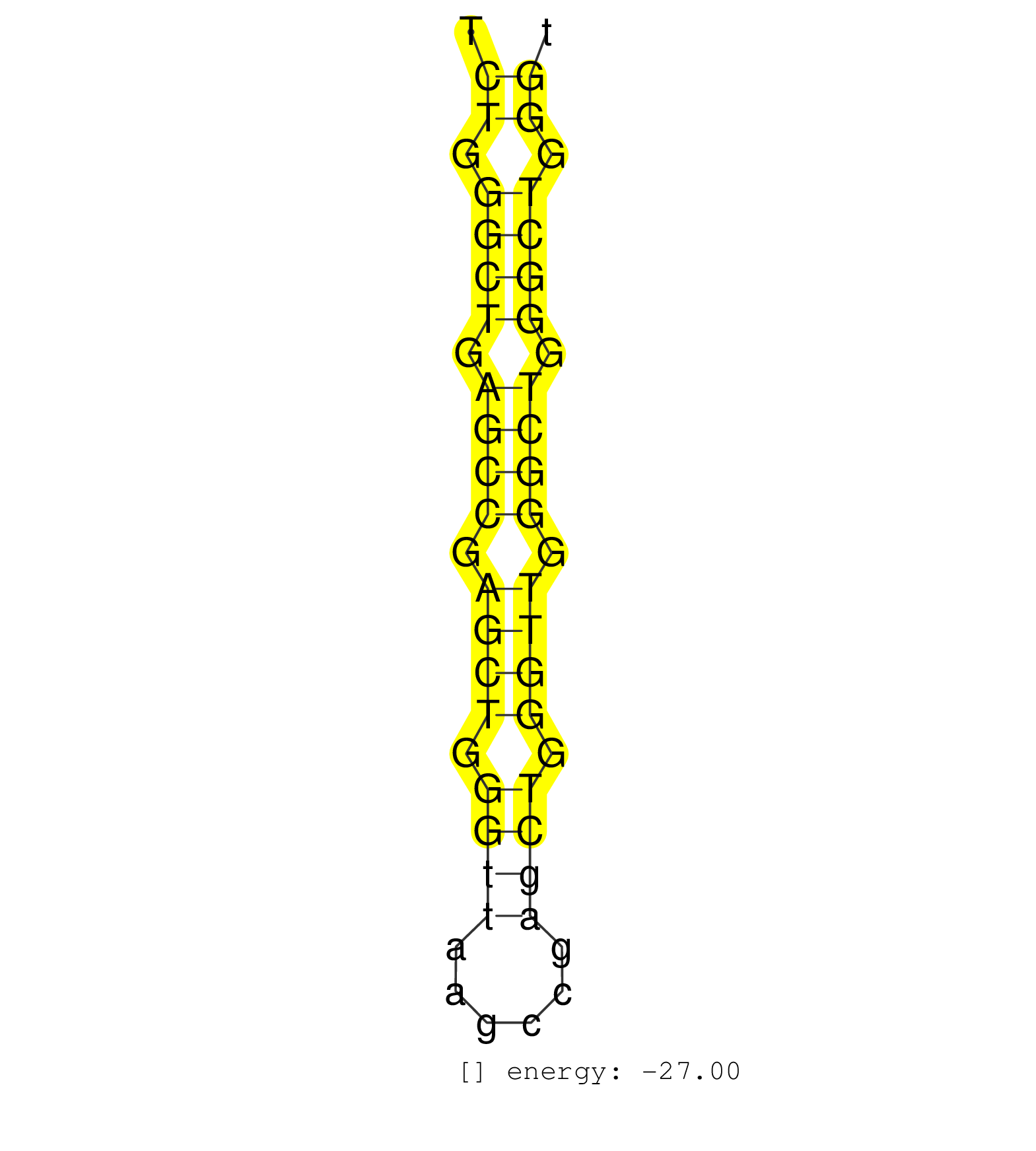
| GCTGAACTGGGCTGGGTTGAGCTGGGCTGGGCTGAGTTGAGCCAGGCTGATCTGGGCTGAGCCGAGCTGGGTTAAGCCGAGCTGGGTTGGGCTGGGCTGGGTTGGGCTGGGCTGAGCCGGACTGGGTTGGGCTGAGCTGAGCTGGACTGGGC ...................................................((.((((.((((.((((.((((......)))).)))).)))).)))).))................................................... ..................................................51.................................................102................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR015362(GSM380327) Naïve B Cell (Naive138). (B cell) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR015365(GSM380330) Memory B cells (MM139). (B cell) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191438(GSM715548) 175genomic small RNA (size selected RNA from . (breast) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR015361(GSM380326) Memory B cells (MM55). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR040037(GSM532922) G243T. (cervix) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR015358(GSM380323) Naïve B Cell (Naive39). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR191501(GSM715611) 8genomic small RNA (size selected RNA from to. (breast) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR191553(GSM715663) 96genomic small RNA (size selected RNA from t. (breast) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM339994(GSM339994) hues6. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR191529(GSM715639) 131genomic small RNA (size selected RNA from . (breast) | TAX577579(Rovira) total RNA. (breast) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................CTGGGTTGGGCTGGGCTGGG................................................... | 20 | 3 | 14.00 | 14.00 | 7.33 | 1.67 | 1.67 | 1.67 | 0.33 | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTa................................................. | 22 | A | 10.00 | 1.00 | 2.50 | 3.50 | 0.50 | 0.50 | - | - | - | - | - | 1.00 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTGGGCTGGGCTGGGTTGGG.............................................. | 20 | 2 | 10.00 | 10.00 | 8.00 | 0.50 | - | - | 1.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGGCaa...................................... | 23 | AA | 9.00 | 0.00 | 1.00 | 2.00 | 1.00 | 1.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTT................................................. | 22 | 1 | 6.00 | 6.00 | 1.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTa..................................................... | 18 | A | 5.33 | 0.00 | 0.67 | 1.33 | 0.67 | 0.67 | - | - | 0.67 | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTTa................................................ | 23 | A | 4.50 | 6.00 | 4.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTaa................................................ | 23 | AA | 4.50 | 1.00 | 3.00 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GCTGGGCTGAGCCGGgct............................. | 18 | GCT | 4.00 | 0.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTTG................................................ | 23 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGGGTTGGGCTGGGCTGGGTT................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTgaa............................................... | 24 | GAA | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGGCTG...................................... | 23 | 2 | 1.50 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGGGTTGGGCTGGGCTGGGTa................................................. | 21 | A | 1.50 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGGGTTGGGCTGGGCTGGG................................................... | 19 | 5 | 1.40 | 1.40 | 0.80 | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | 0.20 | - | - | - | - |
| ................................................................................................CTGGGTTGGGCTGGGCaaa..................................... | 19 | AAA | 1.33 | 0.00 | - | - | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGaa................................................. | 22 | AA | 1.33 | 14.00 | 1.00 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TTGGGCTGAGCTGAGata........ | 18 | ATA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................GCTGGGTTGGGCTGGGCTGGGTTG................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGCCGAGCTGGGTTAAGCCGAGCTGGGTTG............................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGGCTGGGCTGGGTTGtatt............................................ | 20 | TATT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CGGACTGGGTTGGGCTGAGCTGAGCTctg...... | 29 | CTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGGTTGGGCTGGGCTGGGTTGGGCTGGGCTG...................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CGGACTGGGTTGGGCTctg................ | 19 | CTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TTGGGCTGAGCTGAGacc........ | 18 | ACC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GGCTGGGTTGGGCTGGGCTGAGCCGGA............................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTGAGCTGAGCTGGACTGGGCtgaa | 25 | TGAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTGGGCTGGGCTGGGTTGGGCT............................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGT.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GAGCTGAGCTGGACTGGGCtgaa | 23 | TGAA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGGCTa...................................... | 23 | A | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GGCTGAGCCGGACTGGGTTG....................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGGCTaa..................................... | 24 | AA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................ATCTGGGCTGAGCCGAGCTGGG................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGAGCTGGGCTGGGCTGAGca.................................................................................................................. | 21 | CA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AGCTGAGCTGGACTGGGCtgat | 22 | TGAT | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTGGGCTGGGCTGGGTTGGGta............................................ | 22 | TA | 1.00 | 10.00 | 0.33 | - | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GCTGGGCTGAGCCGGACTGaa.......................... | 21 | AA | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AGCCGGACTGGGTTGtgt.................... | 18 | TGT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TTGGGCTGGGCTGAGCtctg............................... | 20 | TCTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGGta....................................... | 22 | TA | 0.86 | 0.43 | 0.71 | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGGTTGGGCTGGGCTaaaa.................................................. | 19 | AAAA | 0.67 | 0.00 | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGcgta................................................. | 22 | CGTA | 0.67 | 0.00 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGGGTTGGGCTGGGCTGtg................................................... | 19 | TG | 0.67 | 0.00 | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGG.................................................... | 19 | 5 | 0.60 | 0.60 | - | 0.20 | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGGtat...................................... | 23 | TAT | 0.57 | 0.43 | - | - | 0.29 | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGtg......................................... | 20 | TG | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GGCTGGGTTGGGCTGGGCTG...................................... | 20 | 4 | 0.50 | 0.50 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - |
| ..................................................................................TGGGTTGGGCTGGGCTGGGTTa................................................ | 22 | A | 0.50 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................GAGCCGAGCTGGGTTgatt........................................................................... | 19 | GATT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................CTGGGCTGAGCCGAGCTact................................................................................. | 20 | ACT | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......CTGGGCTGGGTTGAGggtc............................................................................................................................... | 19 | GGTC | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTaatt.............................................. | 25 | AATT | 0.50 | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTaaa............................................... | 24 | AAA | 0.50 | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GTTGGGCTGGGCTGGaaga................................................ | 19 | AAGA | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................GGCTGGGCTGAGTTGAGCCAGGC......................................................................................................... | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GGCTGGGTTGGGCTGGctgg...................................... | 20 | CTGG | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GAGCTGGGCTGGGCTGAGTTGAGCtctg.......................................................................................................... | 28 | TCTG | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................GCTGGGCTGGGCTGAGaaaa................................................................................................................ | 20 | AAAA | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGTTt................................................ | 23 | T | 0.50 | 6.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....ACTGGGCTGGGTTGAGagg................................................................................................................................ | 19 | AGG | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGG......................................... | 20 | 7 | 0.43 | 0.43 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGGtta...................................... | 23 | TTA | 0.43 | 0.43 | - | - | - | - | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| ..................................................................................TGGGTTGGGCTGGGCTGGGcaa................................................ | 22 | CAA | 0.40 | 1.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - |
| .................................................................................CTGGGTTGGGCTGGGCTGGGag................................................. | 22 | AG | 0.33 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGGGCTGGGCTGGGTgaa............................................... | 18 | GAA | 0.33 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - |
| ...........CTGGGTTGAGCTGGGCTGG.......................................................................................................................... | 19 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| .....................CTGGGCTGGGCTGAGaaa................................................................................................................. | 18 | AAA | 0.33 | 0.00 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTGGGCTGGGCTGGGTcggg.............................................. | 20 | CGGG | 0.33 | 0.00 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CTGGGCTGGGTTGGGCTGGGaaa...................................... | 23 | AAA | 0.14 | 0.43 | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGGGTTGGGCTGGGCTGG.................................................... | 18 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| GCTGAACTGGGCTGGGTTGAGCTGGGCTGGGCTGAGTTGAGCCAGGCTGATCTGGGCTGAGCCGAGCTGGGTTAAGCCGAGCTGGGTTGGGCTGGGCTGGGTTGGGCTGGGCTGAGCCGGACTGGGTTGGGCTGAGCTGAGCTGGACTGGGC ...................................................((.((((.((((.((((.((((......)))).)))).)))).)))).))................................................... ..................................................51.................................................102................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR015362(GSM380327) Naïve B Cell (Naive138). (B cell) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR015365(GSM380330) Memory B cells (MM139). (B cell) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191438(GSM715548) 175genomic small RNA (size selected RNA from . (breast) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR015361(GSM380326) Memory B cells (MM55). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR040037(GSM532922) G243T. (cervix) | SRR139208(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) | SRR015358(GSM380323) Naïve B Cell (Naive39). (B cell) | TAX577739(Rovira) total RNA. (breast) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR191501(GSM715611) 8genomic small RNA (size selected RNA from to. (breast) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR191553(GSM715663) 96genomic small RNA (size selected RNA from t. (breast) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM339994(GSM339994) hues6. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR191529(GSM715639) 131genomic small RNA (size selected RNA from . (breast) | TAX577579(Rovira) total RNA. (breast) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) |
|---|