| (1) AGO2.ip | (2) B-CELL | (12) BRAIN | (7) BREAST | (17) CELL-LINE | (2) CERVIX | (2) HEART | (6) HELA | (6) OTHER | (10) SKIN | (2) UTERUS |
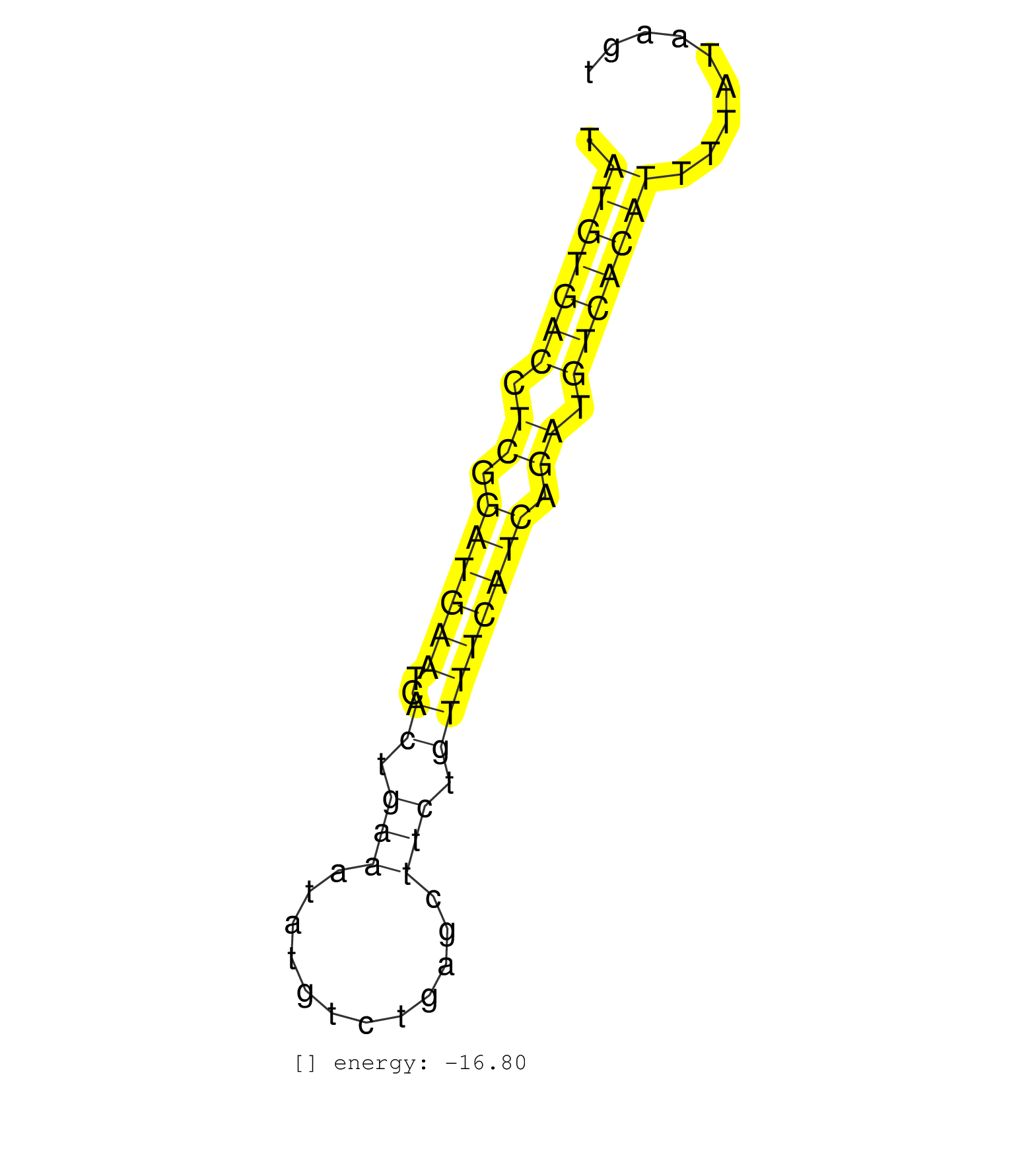
| GTGCATTGGAATTAAATGGAGCTGGGTCTGAATTCCAGTCTATCACTTGTTATGTGACCTCGGATGAATCACTGAAATATGTCTGAGCTTCTGTTTCATCAGATGTCACATTTTATAAGTGATAGTGCTATTGTGAGGATTAAATAAGATAGTACCTGTCAAAT ...................................................(((((((.((.((((((..((.(((............))).)))))))).)).)))))))........................................................... ..................................................51...................................................................120................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189782 | SRR189786 | SRR189787 | SRR037937(GSM510475) 293cand2. (cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR189783 | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR189784 | SRR037938(GSM510476) 293Red. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037941(GSM510479) 293DroshaTN. (cell line) | TAX577579(Rovira) total RNA. (breast) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | GSM416733(GSM416733) HEK293. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR037931(GSM510469) 293GFP. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029124(GSM416753) HeLa. (hela) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038858(GSM458541) MEL202. (cell line) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577745(Rovira) total RNA. (breast) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040030(GSM532915) G013N. (cervix) | SRR037935(GSM510473) 293cand3. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR139170(SRX050649) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR189785 | TAX577741(Rovira) total RNA. (breast) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR191415(GSM715525) 33genomic small RNA (size selected RNA from t. (breast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040012(GSM532897) G648N. (cervix) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | DRR001485(DRX001039) Hela long total cell fraction, LNA(+). (hela) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | DRR001487(DRX001041) Hela long nuclear cell fraction, LNA(+). (hela) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................TTTCATCAGATGTCACATTTTAT................................................ | 23 | 1 | 125.00 | 125.00 | 65.00 | 14.00 | 5.00 | 14.00 | - | - | - | - | 2.00 | - | 5.00 | 2.00 | 2.00 | - | - | 6.00 | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATCA............................................................................................. | 21 | 1 | 72.00 | 72.00 | 4.00 | 2.00 | - | - | 9.00 | 3.00 | 5.00 | 2.00 | - | 2.00 | 1.00 | - | - | 7.00 | 2.00 | - | 6.00 | - | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | 3.00 | - | 1.00 | 1.00 | - | 1.00 | 2.00 | 1.00 | 2.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 |
| .............................................................................................TTTCATCAGATGTCACATTTT.................................................. | 21 | 1 | 38.00 | 38.00 | 30.00 | - | 1.00 | - | - | - | - | - | - | - | - | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATC.............................................................................................. | 20 | 1 | 34.00 | 34.00 | - | - | - | - | - | 10.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 4.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - |
| .............................................................................................TTTCATCAGATGTCACATTTTA................................................. | 22 | 1 | 29.00 | 29.00 | 25.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAAT............................................................................................... | 19 | 1 | 28.00 | 28.00 | - | - | - | - | 7.00 | 2.00 | 6.00 | - | - | 4.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACAT..................................................... | 18 | 1 | 26.00 | 26.00 | - | - | - | 14.00 | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTctat................................................ | 23 | CTAT | 24.00 | 0.00 | - | 14.00 | 9.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTcat................................................ | 23 | CAT | 13.00 | 1.00 | 1.00 | 7.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTat................................................. | 22 | AT | 12.00 | 1.00 | 3.00 | 5.00 | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTTAc................................................ | 23 | C | 11.00 | 29.00 | - | 8.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAA................................................................................................ | 18 | 1 | 9.00 | 9.00 | - | - | - | - | 3.00 | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGA................................................................................................. | 17 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATCAC............................................................................................ | 22 | 1 | 6.00 | 6.00 | - | - | - | - | - | 2.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TTATGTGACCTCGGATGAATC.............................................................................................. | 21 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAAa............................................................................................... | 19 | A | 4.00 | 9.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATCACT........................................................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTcta................................................. | 22 | CTA | 3.00 | 0.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTTt................................................. | 22 | T | 3.00 | 38.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATCt............................................................................................. | 21 | T | 2.00 | 34.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................ATGTGACCTCGGATGAATCACTG.......................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TTATGTGACCTCGGATGAAT............................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTTATAA.............................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATctta................................................. | 22 | CTTA | 2.00 | 26.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGTTTCATCAGATGTCACATTTT.................................................. | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATta............................................................................................. | 21 | TA | 2.00 | 28.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTatt................................................ | 23 | ATT | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTa................................................... | 20 | A | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAAccac............................................................................................ | 22 | CCAC | 1.00 | 9.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAAcca............................................................................................. | 21 | CCA | 1.00 | 9.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATctat................................................. | 22 | CTAT | 1.00 | 26.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTgtag................................................ | 23 | GTAG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTTATt............................................... | 24 | T | 1.00 | 125.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTat.................................................. | 21 | AT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACAccat.................................................. | 21 | CCAT | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTgtt................................................. | 22 | GTT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATCAt............................................................................................ | 22 | T | 1.00 | 72.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTTATtt.............................................. | 25 | TT | 1.00 | 125.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTccac................................................ | 23 | CCAC | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTTAct............................................... | 24 | CT | 1.00 | 29.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................ATGTGACCTCGGATGAATCAC............................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTca................................................. | 22 | CA | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTcac................................................ | 23 | CAC | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATCACTG.......................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTT................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTa.................................................. | 21 | A | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACgtat................................................... | 20 | GTAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATctt.................................................. | 21 | CTT | 1.00 | 26.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TTATGTGACCTCGGATGAAa............................................................................................... | 20 | A | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATGTGACCTCGGATGAATCc............................................................................................. | 21 | C | 1.00 | 34.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTTATAt.............................................. | 25 | T | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTTCATCAGATGTCACATTTTATA............................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............AAATGGAGCTGGGTCTGAATTCtg............................................................................................................................... | 24 | TG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTGCATTGGAATTAAATGGAGCTGGGTCTGAATTCCAGTCTATCACTTGTTATGTGACCTCGGATGAATCACTGAAATATGTCTGAGCTTCTGTTTCATCAGATGTCACATTTTATAAGTGATAGTGCTATTGTGAGGATTAAATAAGATAGTACCTGTCAAAT ...................................................(((((((.((.((((((..((.(((............))).)))))))).)).)))))))........................................................... ..................................................51...................................................................120................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR189782 | SRR189786 | SRR189787 | SRR037937(GSM510475) 293cand2. (cell line) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR189783 | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR189784 | SRR037938(GSM510476) 293Red. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037941(GSM510479) 293DroshaTN. (cell line) | TAX577579(Rovira) total RNA. (breast) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | GSM416733(GSM416733) HEK293. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR037931(GSM510469) 293GFP. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029124(GSM416753) HeLa. (hela) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038858(GSM458541) MEL202. (cell line) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577745(Rovira) total RNA. (breast) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040030(GSM532915) G013N. (cervix) | SRR037935(GSM510473) 293cand3. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR139170(SRX050649) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR189785 | TAX577741(Rovira) total RNA. (breast) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR191415(GSM715525) 33genomic small RNA (size selected RNA from t. (breast) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040012(GSM532897) G648N. (cervix) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | DRR001485(DRX001039) Hela long total cell fraction, LNA(+). (hela) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | DRR001487(DRX001041) Hela long nuclear cell fraction, LNA(+). (hela) |
|---|