| (6) B-CELL | (5) BREAST | (5) CELL-LINE | (2) CERVIX | (1) HEART | (8) LIVER | (1) OTHER | (1) RRP40.ip | (4) SKIN | (2) UTERUS | (1) XRN.ip |
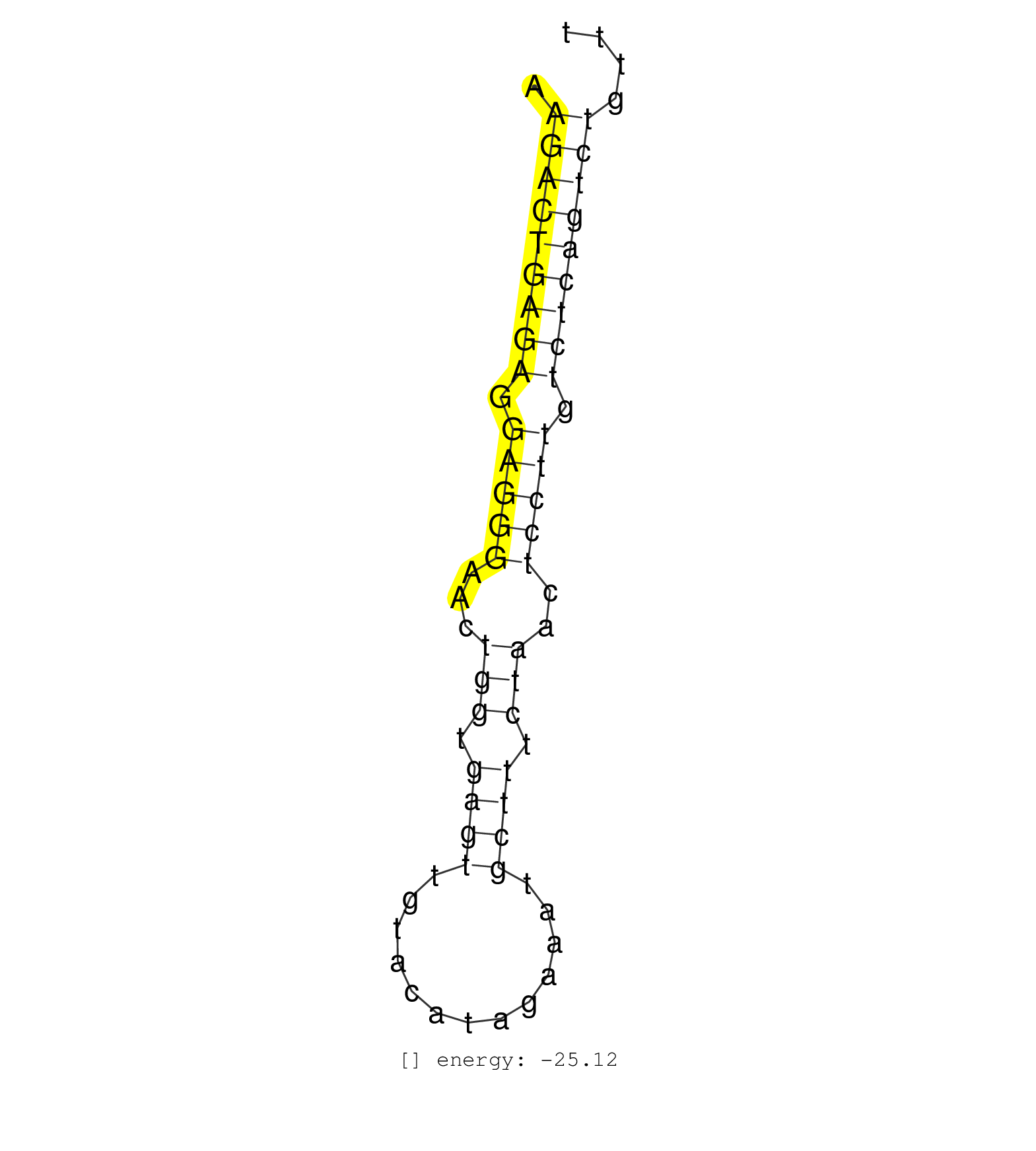
| CTGGGGGTCACCATCACTCAGTGAAAGTGTGAGCAGGAAAAATCGCAGAGAAGACTGAGAGGAGGGAACTGGTGAGTTGTACATAGAAATGCTTTCTAACTCCTTGTCTCAGTCTGTTTTTGCTACCAGTGAGGCTTACAATGTTTGAAAGACCCCAATCCCAGCATGC ...................................................(((((((((.(((((...(((.((((.............)))).)))..))))).)))))))))...................................................... ..................................................51..................................................................119................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | TAX577580(Rovira) total RNA. (breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | GSM532880(GSM532880) G659T. (cervix) | TAX577742(Rovira) total RNA. (breast) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR444044(SRX128892) Sample 5cDNABarcode: AF-PP-340: ACG CTC TTC C. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR040031(GSM532916) G013T. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR038854(GSM458537) MM653. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR191560(GSM715670) 77genomic small RNA (size selected RNA from t. (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | TAX577453(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................AAGACTGAGAGGAGGGAA..................................................................................................... | 18 | 4 | 5.75 | 5.75 | - | 0.75 | 1.25 | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | 0.25 | - | 0.25 | 0.25 | 0.25 | - | 0.25 | - | - | 0.25 | 0.25 | 0.25 | 0.25 | - | 0.25 | - | 0.25 | - |
| ...............................................GAGAAGACTGAGAGGAGGGAA..................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGAAGACTGAGAGGAGGGAACT................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................GTCTCAGTCTGTTTTTtt.............................................. | 18 | TT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGGAGGGAACTGGTGAGTTGTA........................................................................................ | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................AAGACTGAGAGGAGGGgaag................................................................................................... | 20 | GAAG | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................GGAAAAATCGCAGAGggcc................................................................................................................... | 19 | GGCC | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................AGAGAAGACTGAGAGcgg......................................................................................................... | 18 | CGG | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................GCAGAGAAGACTGAGAGG........................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................AAGACTGAGAGGAGGGA...................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGGGGGTCACCATCACTCAGTGAAAGTGTGAGCAGGAAAAATCGCAGAGAAGACTGAGAGGAGGGAACTGGTGAGTTGTACATAGAAATGCTTTCTAACTCCTTGTCTCAGTCTGTTTTTGCTACCAGTGAGGCTTACAATGTTTGAAAGACCCCAATCCCAGCATGC ...................................................(((((((((.(((((...(((.((((.............)))).)))..))))).)))))))))...................................................... ..................................................51..................................................................119................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | TAX577580(Rovira) total RNA. (breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | GSM532880(GSM532880) G659T. (cervix) | TAX577742(Rovira) total RNA. (breast) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR189778(GSM714638) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR444044(SRX128892) Sample 5cDNABarcode: AF-PP-340: ACG CTC TTC C. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR040031(GSM532916) G013T. (cervix) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR038854(GSM458537) MM653. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR191560(GSM715670) 77genomic small RNA (size selected RNA from t. (breast) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR033728(GSM497073) MALT (MALT413). (B cell) | TAX577453(Rovira) total RNA. (breast) | TAX577745(Rovira) total RNA. (breast) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgGGGGGTCACCATCACTC...................................................................................................................................................... | 19 | cg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................gtccGTCTCAGTCTGTTTT................................................. | 19 | gtcc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................tacgTCGCAGAGAAGACTG................................................................................................................ | 19 | tacg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GGGAACTGGTGAGTTGTACATAGAAATG.............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| cctGGGGTCACCATCACT....................................................................................................................................................... | 18 | cct | 0.75 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | 0.25 | - | - |
| acctGGGGTCACCATCACT....................................................................................................................................................... | 19 | acct | 0.75 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | 0.25 |
| ..................................................................AACTGGTGAGTTGTACA...................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...GGGGTCACCATCACT....................................................................................................................................................... | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - |
| .ctGGGGTCACCATCACT....................................................................................................................................................... | 17 | ct | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |