| (1) AGO2.ip | (1) B-CELL | (3) BREAST | (4) CELL-LINE | (1) CERVIX | (2) HEART | (2) HELA | (1) LIVER | (2) OTHER | (4) SKIN | (1) TESTES |
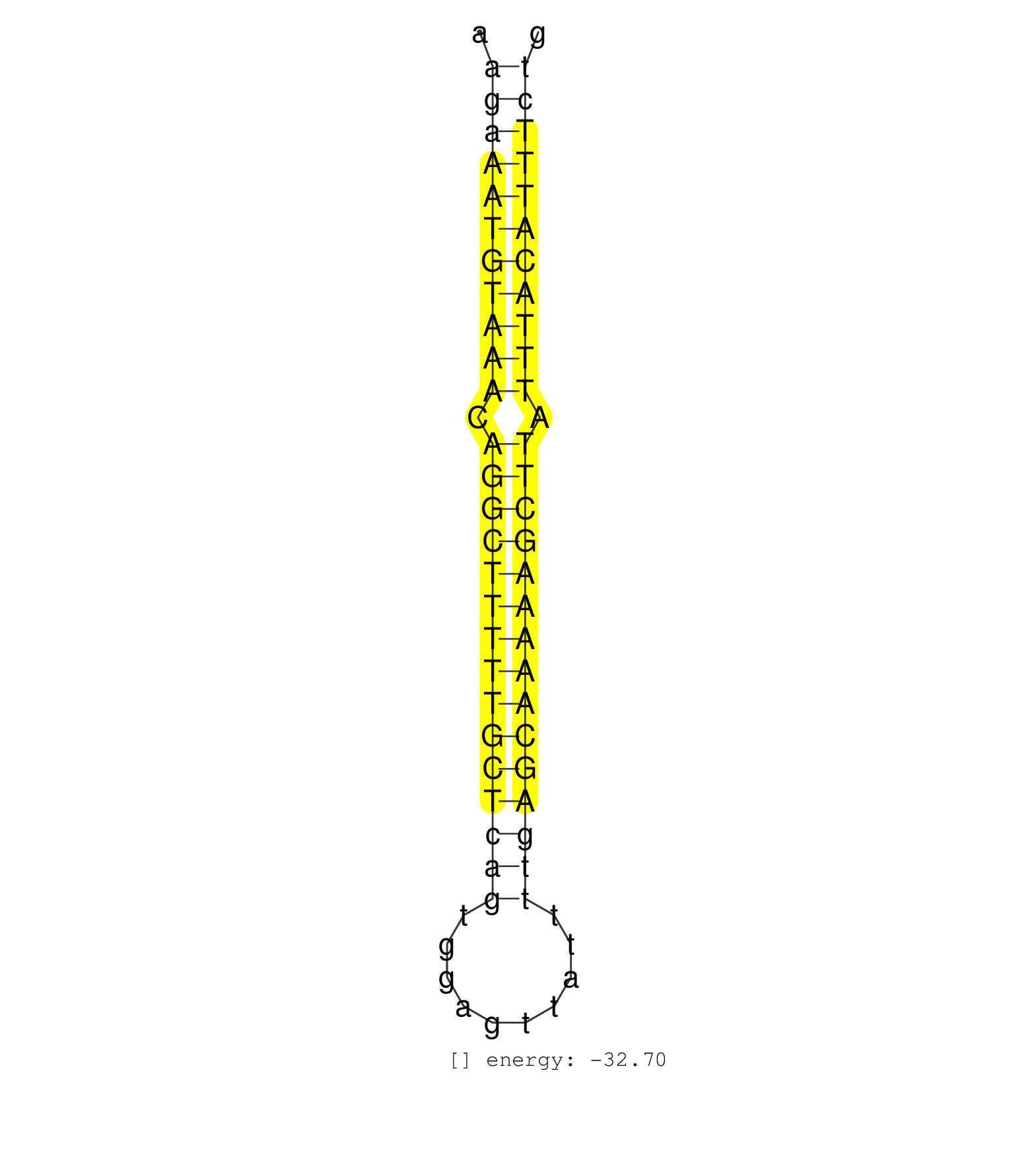
| TGTGATGGGTGATATTTTTTTCTCATTTTACAGATGAGAAAACTGAGGCTAAGAAATGTAAACAGGCTTTTTGCTCAGTGGAGTTATTTTGAGCAAAAAGCTTATTTACATTTCTGAGTCTCAGTTTCCCCAACAAGTAGCTGTTATTTTGATAACCCAGGAATAC ...................................................(((((((((((.(((((((((((((((..........))))))))))))))).)))))))))))................................................... ..................................................51...............................................................116................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR029129(GSM416758) SW480. (cell line) | SRR139170(SRX050649) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR189784 | SRR191606(GSM715716) 84genomic small RNA (size selected RNA from t. (breast) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR040007(GSM532892) G601T. (cervix) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR189786 | SRR191422(GSM715532) 129genomic small RNA (size selected RNA from . (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139210(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................AATGTAAACAGGCTTTTTGCT........................................................................................... | 21 | 1 | 14.00 | 14.00 | 13.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AATGTAAACAGGCTTTTTG............................................................................................. | 19 | 1 | 8.00 | 8.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................AGCAAAAAGCTTATTTACATTT..................................................... | 22 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGAGTCTCAGTTTCCCCAggta.............................. | 22 | GGTA | 3.00 | 0.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................ATGTAAACAGGCTTTTTGCTCA......................................................................................... | 22 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGAGTCTCAGTTTCCCCAggtt.............................. | 22 | GGTT | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................TGAGTCTCAGTTTCCCCcggt............................... | 21 | CGGT | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................TGTAAACAGGCTTTTTGCTCAGa....................................................................................... | 23 | A | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TGTAAACAGGCTTTTTGCTCAGT....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................ATGTAAACAGGCTTTTTGagac......................................................................................... | 22 | AGAC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................TTACATTTCTGAGTCca............................................ | 17 | CA | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............ATTTTTTTCTCATTTTAtttt.................................................................................................................................... | 21 | TTTT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................AATGTAAACAGGCTTTTTt............................................................................................. | 19 | T | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AATGTAAACAGGCTTTTTGC............................................................................................ | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................ATGTAAACAGGCTTTTTGCT........................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TTACAGATGAGAAAACTGAGttgc................................................................................................................... | 24 | TTGC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGAGTCTCAGTTTCCCCAttta.............................. | 22 | TTTA | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................ATGTAAACAGGCTTTTTGCTta......................................................................................... | 22 | TA | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AATGTAAACAGGCTTctt.............................................................................................. | 18 | CTT | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AATGTAAACAGGCTTTTTGCTC.......................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TGTAAACAGGCTTTTTGCTCA......................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AATGTAAACAGGCTTTTTa............................................................................................. | 19 | A | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CAAAAAGCTTATTTACATTTC.................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GCAAAAAGCTTATTTACATTTC.................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................AATGTAAACAGGCTTTatg............................................................................................. | 19 | ATG | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGTGATGGGTGATATTTTTTTCTCATTTTACAGATGAGAAAACTGAGGCTAAGAAATGTAAACAGGCTTTTTGCTCAGTGGAGTTATTTTGAGCAAAAAGCTTATTTACATTTCTGAGTCTCAGTTTCCCCAACAAGTAGCTGTTATTTTGATAACCCAGGAATAC ...................................................(((((((((((.(((((((((((((((..........))))))))))))))).)))))))))))................................................... ..................................................51...............................................................116................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR029129(GSM416758) SW480. (cell line) | SRR139170(SRX050649) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR189784 | SRR191606(GSM715716) 84genomic small RNA (size selected RNA from t. (breast) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR040007(GSM532892) G601T. (cervix) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR189786 | SRR191422(GSM715532) 129genomic small RNA (size selected RNA from . (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | TAX577743(Rovira) total RNA. (breast) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR139210(SRX050635) FLASHPage purified small RNA (~15-40nt) from . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................actgTGAGTCTCAGTTTCCCCA.................................. | 22 | actg | 3.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................caagAGTGGAGTTATTTTG........................................................................... | 19 | caag | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................ccggATTTTACAGATGAGAA.............................................................................................................................. | 20 | ccgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................acaTTGAGCAAAAAGCTT............................................................... | 18 | aca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................atggCTCATTTTACAGATGAG................................................................................................................................ | 21 | atgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................acTTTGAGCAAAAAGCTT............................................................... | 18 | ac | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGATATTTTTTTCTCATT........................................................................................................................................... | 18 | 4 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |