| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (9) B-CELL | (13) BRAIN | (7) BREAST | (18) CELL-LINE | (2) CERVIX | (2) HEART | (3) HELA | (15) LIVER | (2) RRP40.ip | (7) SKIN | (3) UTERUS | (1) XRN.ip |
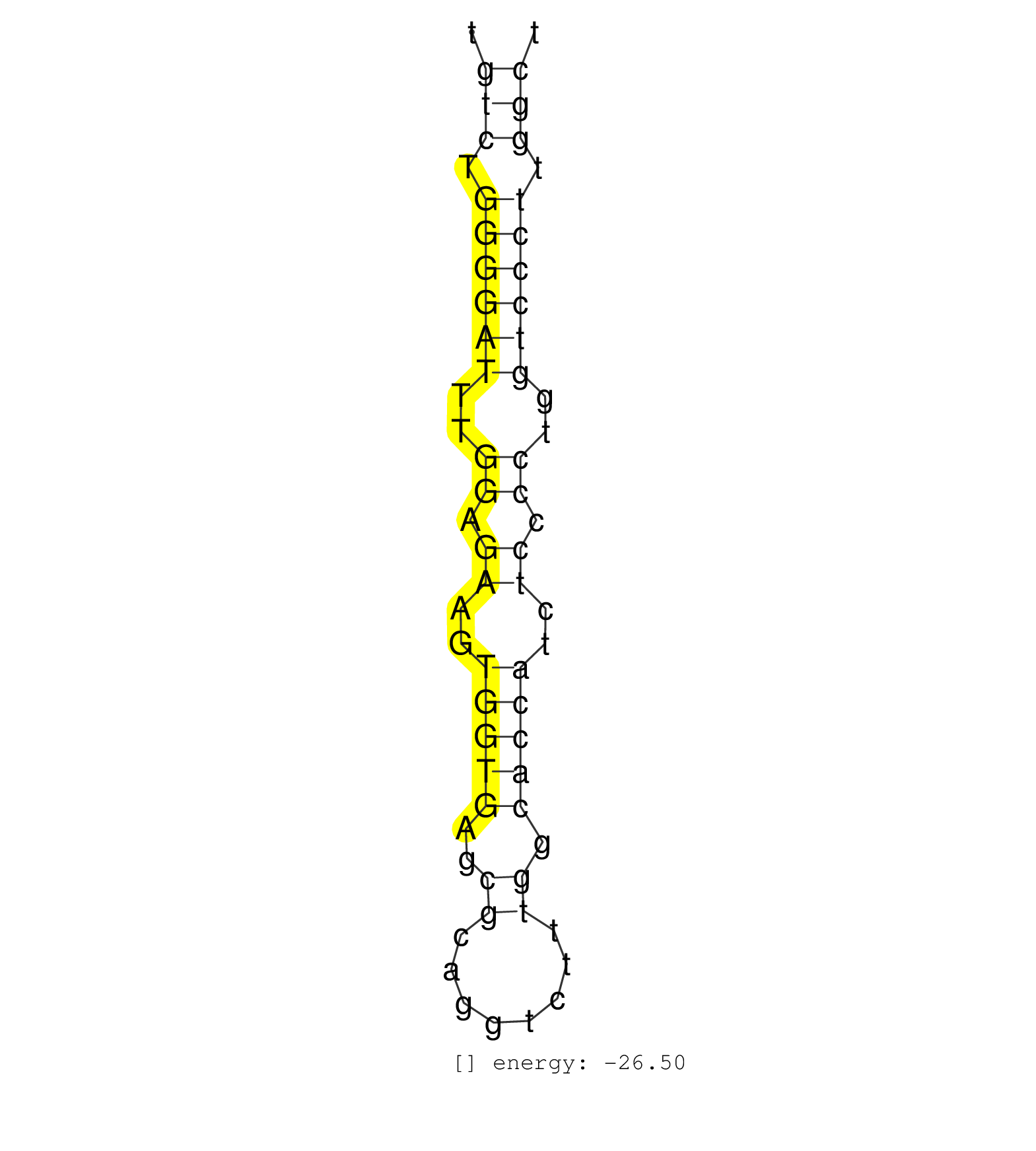
| TGCTCAGATGTCATCTTTTTTCTCCACTGCCCAGCTGTCTTGCTAAACAGTGTCTGGGGATTTGGAGAAGTGGTGAGCGCAGGTCTTTGGCACCATCTCCCCTGGTCCCTTGGCTCTGTCCATAGCCTTCCCAGCCACTGCTGGCCTCCAACGCTGAATCCTGCA ...................................................(((.((((((..((.((..(((((..((........)).)))))..)).))..)))))).)))................................................... ..................................................51..............................................................115................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR038861(GSM458544) MM466. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR029128(GSM416757) H520. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR038862(GSM458545) MM472. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207117(GSM721079) Whole cell RNA. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR038854(GSM458537) MM653. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR038855(GSM458538) D10. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577453(Rovira) total RNA. (breast) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM532876(GSM532876) G547T. (cervix) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577580(Rovira) total RNA. (breast) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR040031(GSM532916) G013T. (cervix) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191443(GSM715553) 108genomic small RNA (size selected RNA from . (breast) | TAX577589(Rovira) total RNA. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | DRR001485(DRX001039) Hela long total cell fraction, LNA(+). (hela) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR191571(GSM715681) 63genomic small RNA (size selected RNA from t. (breast) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................TGGGGATTTGGAGAAGTGGTGA......................................................................................... | 22 | 1 | 628.00 | 628.00 | 123.00 | 166.00 | 54.00 | 1.00 | 26.00 | 14.00 | 25.00 | 15.00 | 12.00 | 19.00 | 12.00 | 10.00 | 7.00 | - | 6.00 | 8.00 | 7.00 | 8.00 | 2.00 | 9.00 | 9.00 | 6.00 | 6.00 | 2.00 | 7.00 | 6.00 | 5.00 | 4.00 | 4.00 | 6.00 | - | 5.00 | 4.00 | 4.00 | 2.00 | 2.00 | 2.00 | 2.00 | - | 3.00 | 1.00 | 2.00 | 3.00 | 2.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | 2.00 | - | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - |
| ......................................................TGGGGATTTGGAGAAGTGGTG.......................................................................................... | 21 | 1 | 213.00 | 213.00 | 65.00 | 29.00 | 17.00 | 11.00 | 6.00 | 13.00 | 7.00 | 6.00 | 7.00 | 1.00 | 2.00 | 3.00 | 2.00 | 2.00 | 1.00 | 1.00 | 3.00 | 2.00 | - | - | 1.00 | 3.00 | 2.00 | 4.00 | 1.00 | 2.00 | 2.00 | 2.00 | 1.00 | - | 6.00 | - | 1.00 | - | - | 2.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGG............................................................................................ | 19 | 1 | 50.00 | 50.00 | 3.00 | 1.00 | 1.00 | 23.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | 8.00 | - | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGT........................................................................................... | 20 | 1 | 46.00 | 46.00 | 10.00 | 11.00 | 4.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAa........................................................................................ | 23 | A | 33.00 | 628.00 | 8.00 | 6.00 | 4.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGt......................................................................................... | 22 | T | 26.00 | 213.00 | 9.00 | 7.00 | 1.00 | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAaa....................................................................................... | 24 | AA | 23.00 | 628.00 | 11.00 | 1.00 | - | - | - | - | - | 2.00 | - | 1.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAt........................................................................................ | 23 | T | 17.00 | 628.00 | 2.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGgga......................................................................................... | 22 | GGA | 8.00 | 50.00 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTG............................................................................................. | 18 | 2 | 7.50 | 7.50 | - | - | - | 4.50 | - | - | - | 0.50 | - | - | - | - | - | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................GGGGATTTGGAGAAGTGGTGA......................................................................................... | 21 | 1 | 5.00 | 5.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAat....................................................................................... | 24 | AT | 5.00 | 628.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGgg.......................................................................................... | 21 | GG | 4.00 | 50.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTa.......................................................................................... | 21 | A | 4.00 | 46.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTta......................................................................................... | 22 | TA | 3.00 | 46.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAaaa...................................................................................... | 25 | AAA | 2.00 | 628.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGTGTCTGGGGATTTGGAGgcg............................................................................................... | 22 | GCG | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAtt....................................................................................... | 24 | TT | 2.00 | 628.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGaga......................................................................................... | 22 | AGA | 2.00 | 50.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGaa.......................................................................................... | 21 | AA | 2.00 | 50.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................CTGGGGATTTGGAGAAGTGGTGA......................................................................................... | 23 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGagac........................................................................................ | 23 | AGAC | 1.00 | 50.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGa........................................................................................... | 20 | A | 1.00 | 50.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................GGGGATTTGGAGAAGTGGT........................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................ACGCTGAATCCTGCAgaag | 19 | GAAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGGAGAAGTGGTGAGtag..................................................................................... | 18 | TAG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGttt....................................................................................... | 24 | TTT | 1.00 | 213.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGggaa........................................................................................ | 23 | GGAA | 1.00 | 50.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAacaa..................................................................................... | 26 | ACAA | 1.00 | 628.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTt.......................................................................................... | 21 | T | 1.00 | 46.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGag.......................................................................................... | 21 | AG | 1.00 | 50.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTatt........................................................................................ | 23 | ATT | 1.00 | 46.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............ATCTTTTTTCTCCACTtc....................................................................................................................................... | 18 | TC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TCTGTCCATAGCCTTCCCAGCCACTG......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAaag...................................................................................... | 25 | AAG | 1.00 | 628.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGg........................................................................................... | 20 | G | 1.00 | 50.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................GGGGATTTGGAGAAGgc............................................................................................. | 17 | GC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GGGATTTGGAGAAGTctg........................................................................................... | 18 | CTG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................CTGGGGATTTGGAGAAGTGGTG.......................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGcga......................................................................................... | 22 | CGA | 1.00 | 50.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGcg.......................................................................................... | 21 | CG | 1.00 | 50.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTGAaaaa..................................................................................... | 26 | AAAA | 1.00 | 628.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTaa......................................................................................... | 22 | AA | 1.00 | 46.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGGTat......................................................................................... | 22 | AT | 1.00 | 46.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................CTGGGGATTTGGAGAAGTGGTGt......................................................................................... | 23 | T | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GGGATTTGGAGAAGTGGTG.......................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTGatga......................................................................................... | 22 | ATGA | 0.50 | 7.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGT.............................................................................................. | 17 | 5 | 0.40 | 0.40 | - | - | - | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGTCCATAGCCTTCCCA................................ | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTtg............................................................................................ | 19 | TG | 0.20 | 0.40 | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGGGGATTTGGAGAAGTtgg........................................................................................... | 20 | TGG | 0.20 | 0.40 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................GGATTTGGAGAAGTGG............................................................................................ | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CCACTGCCCAGCTGTC.............................................................................................................................. | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| TGCTCAGATGTCATCTTTTTTCTCCACTGCCCAGCTGTCTTGCTAAACAGTGTCTGGGGATTTGGAGAAGTGGTGAGCGCAGGTCTTTGGCACCATCTCCCCTGGTCCCTTGGCTCTGTCCATAGCCTTCCCAGCCACTGCTGGCCTCCAACGCTGAATCCTGCA ...................................................(((.((((((..((.((..(((((..((........)).)))))..)).))..)))))).)))................................................... ..................................................51..............................................................115................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR038861(GSM458544) MM466. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR029128(GSM416757) H520. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR038862(GSM458545) MM472. (cell line) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207117(GSM721079) Whole cell RNA. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR038854(GSM458537) MM653. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR038855(GSM458538) D10. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577453(Rovira) total RNA. (breast) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM532876(GSM532876) G547T. (cervix) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577580(Rovira) total RNA. (breast) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR040031(GSM532916) G013T. (cervix) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191443(GSM715553) 108genomic small RNA (size selected RNA from . (breast) | TAX577589(Rovira) total RNA. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR038863(GSM458546) MM603. (cell line) | SRR033728(GSM497073) MALT (MALT413). (B cell) | DRR001485(DRX001039) Hela long total cell fraction, LNA(+). (hela) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR191571(GSM715681) 63genomic small RNA (size selected RNA from t. (breast) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................gtcCTCTGTCCATAGCCTTC................................... | 20 | gtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................gggTCTGTCCATAGCCTT.................................... | 18 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................taTTCTCCACTGCCCAGC.................................................................................................................................. | 18 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................taTTGCTAAACAGTGTC............................................................................................................... | 17 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................cgtcCCATAGCCTTCCCAG............................... | 19 | cgtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................CCCTTGGCTCTGTCCA........................................... | 16 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |