| (3) B-CELL | (2) BRAIN | (1) BREAST | (3) CELL-LINE | (1) CERVIX | (2) KIDNEY | (3) LIVER | (2) OTHER | (1) PLACENTA | (3) SKIN |
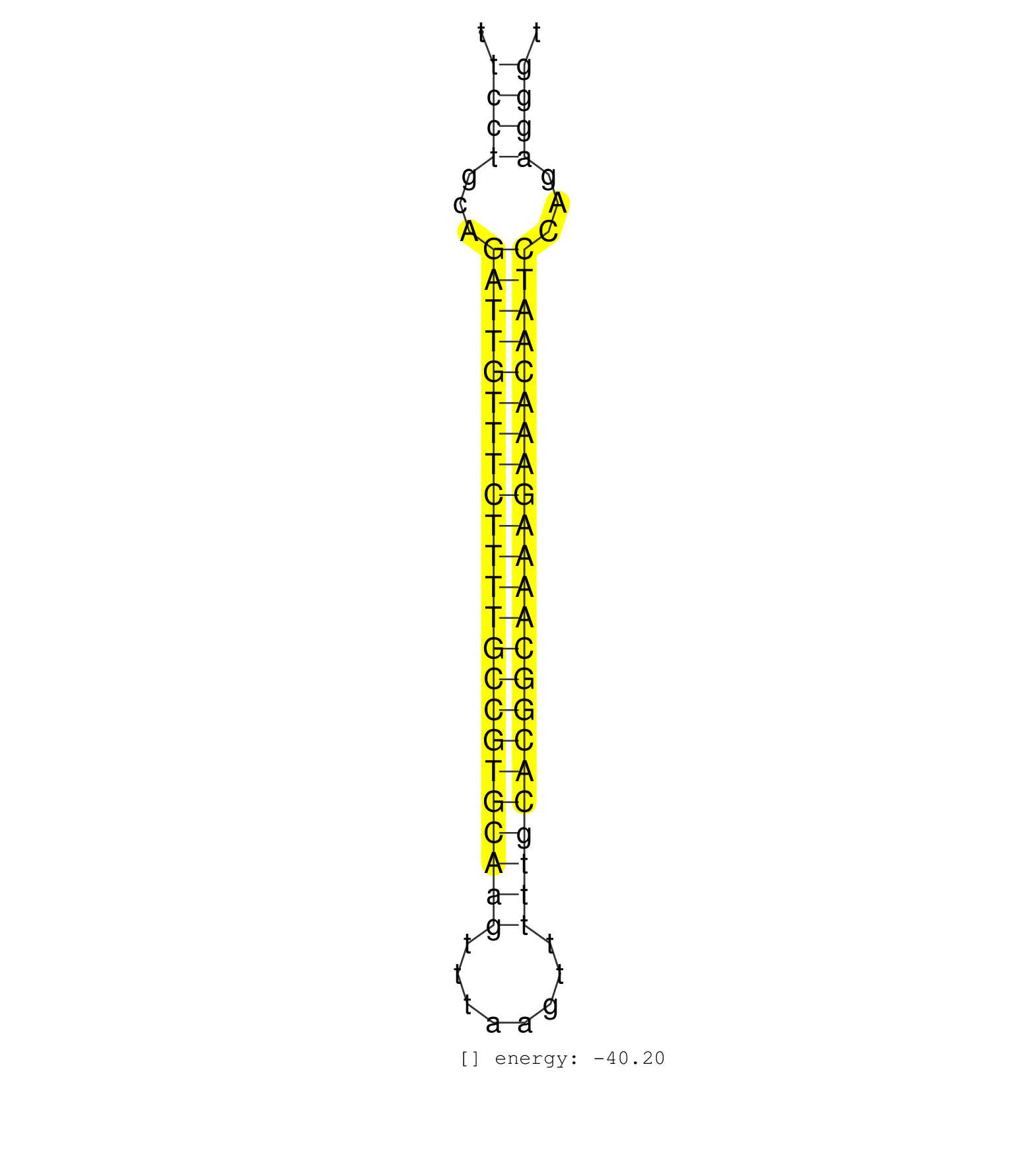
| GACTTAATTAAACTAAATTAAGCACAGCAAAATTTTGTGATTGTCTGTTTTTCCTGCAGATTGTTTCTTTTGCCGTGCAAGTTTAAGTTTTTGCACGGCAAAAGAAACAATCCAGAGGGTAAACAGACAACCCACAAAATGGGAGAAAATCTTTGCGATCTATACATCCG ...................................................((((...(((((((((((((((((((((((........)))))))))))))))))))))))...))))................................................... ..................................................51...................................................................120................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR139172(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR038863(GSM458546) MM603. (cell line) | SRR343334 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR139182(SRX050651) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR040028(GSM532913) G026N. (cervix) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR189782 | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191411(GSM715521) 21genomic small RNA (size selected RNA from t. (breast) | SRR139174(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................AGATTGTTTCTTTTGCCGTGCA........................................................................................... | 22 | 1 | 25.00 | 25.00 | 5.00 | 12.00 | 3.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGCAt.......................................................................................... | 23 | T | 22.00 | 25.00 | 13.00 | 1.00 | 2.00 | 1.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| .............................................................................................CACGGCAAAAGAAACAATCCA........................................................ | 21 | 1 | 21.00 | 21.00 | 3.00 | 8.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CACGGCAAAAGAAACAATCCAG....................................................... | 22 | 1 | 19.00 | 19.00 | 4.00 | 5.00 | 7.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................CACGGCAAAAGAAACAATCC......................................................... | 20 | 1 | 3.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CACGGCAAAAGAAACAATCCAt....................................................... | 22 | T | 2.00 | 21.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CACGGCAAAAGAAACAATa.......................................................... | 19 | A | 1.50 | 0.00 | 1.00 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CACGGCAAAAGAAACAATCCAGt...................................................... | 23 | T | 1.00 | 19.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................GACAACCCACAAAATcg............................ | 17 | CG | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGGGAGAAAATCTTTatc............. | 18 | ATC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGt............................................................................................ | 21 | T | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................ACGGCAAAAGAAACAATCCA........................................................ | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGCAA.......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CACGGCAAAAGAAACAATCCAGA...................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGCAta......................................................................................... | 24 | TA | 1.00 | 25.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TTTAAGTTTTTGCACaaat...................................................................... | 19 | AAAT | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGCAtt......................................................................................... | 24 | TT | 1.00 | 25.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGCct.......................................................................................... | 23 | CT | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................ACCCACAAAATGGGAtacc...................... | 19 | TACC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......TTAAACTAAATTAAGttcc................................................................................................................................................ | 19 | TTCC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGac........................................................................................... | 22 | AC | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGCt........................................................................................... | 22 | T | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................ACGGCAAAAGAAACAATCCAGA...................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................ACGGCAAAAGAAACAATCCAG....................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................CAGATTGTTTCTTTTGCCGTGCAt.......................................................................................... | 24 | T | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................AGATTGTTTCTTTTGCCGTGC............................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CACGGCAAAAGAAACtctg.......................................................... | 19 | TCTG | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .............................................................................................CACGGCAAAAGAAACAATC.......................................................... | 19 | 7 | 0.29 | 0.29 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GACTTAATTAAACTAAATTAAGCACAGCAAAATTTTGTGATTGTCTGTTTTTCCTGCAGATTGTTTCTTTTGCCGTGCAAGTTTAAGTTTTTGCACGGCAAAAGAAACAATCCAGAGGGTAAACAGACAACCCACAAAATGGGAGAAAATCTTTGCGATCTATACATCCG ...................................................((((...(((((((((((((((((((((((........)))))))))))))))))))))))...))))................................................... ..................................................51...................................................................120................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR033710(GSM497055) GCB DLBCL (GCB385). (B cell) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR139172(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR038863(GSM458546) MM603. (cell line) | SRR343334 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR139182(SRX050651) FLASHPage purified small RNA (~15-40nt) from . (liver) | SRR040028(GSM532913) G026N. (cervix) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | SRR139200(SRX050639) FLASHPage purified small RNA (~15-40nt) from . (placenta) | SRR189782 | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191411(GSM715521) 21genomic small RNA (size selected RNA from t. (breast) | SRR139174(SRX050637) FLASHPage purified small RNA (~15-40nt) from . (kidney) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................tATTGTTTCTTTTGCCGTG............................................................................................. | 19 | t | 1.50 | 0.00 | 1.00 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................cgaaCAAGTTTAAGTTTTT.............................................................................. | 19 | cgaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................ACAATCCAGAGGGTAAACAGAC.......................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................cagaGTTTCTTTTGCCGTG............................................................................................. | 19 | caga | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ..........................................................GATTGTTTCTTTTGCCGTG............................................................................................. | 19 | 7 | 0.29 | 0.29 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |