| (4) B-CELL | (3) CELL-LINE | (2) HEART | (1) LUNG | (1) OTHER | (3) SKIN | (1) SPLEEN |
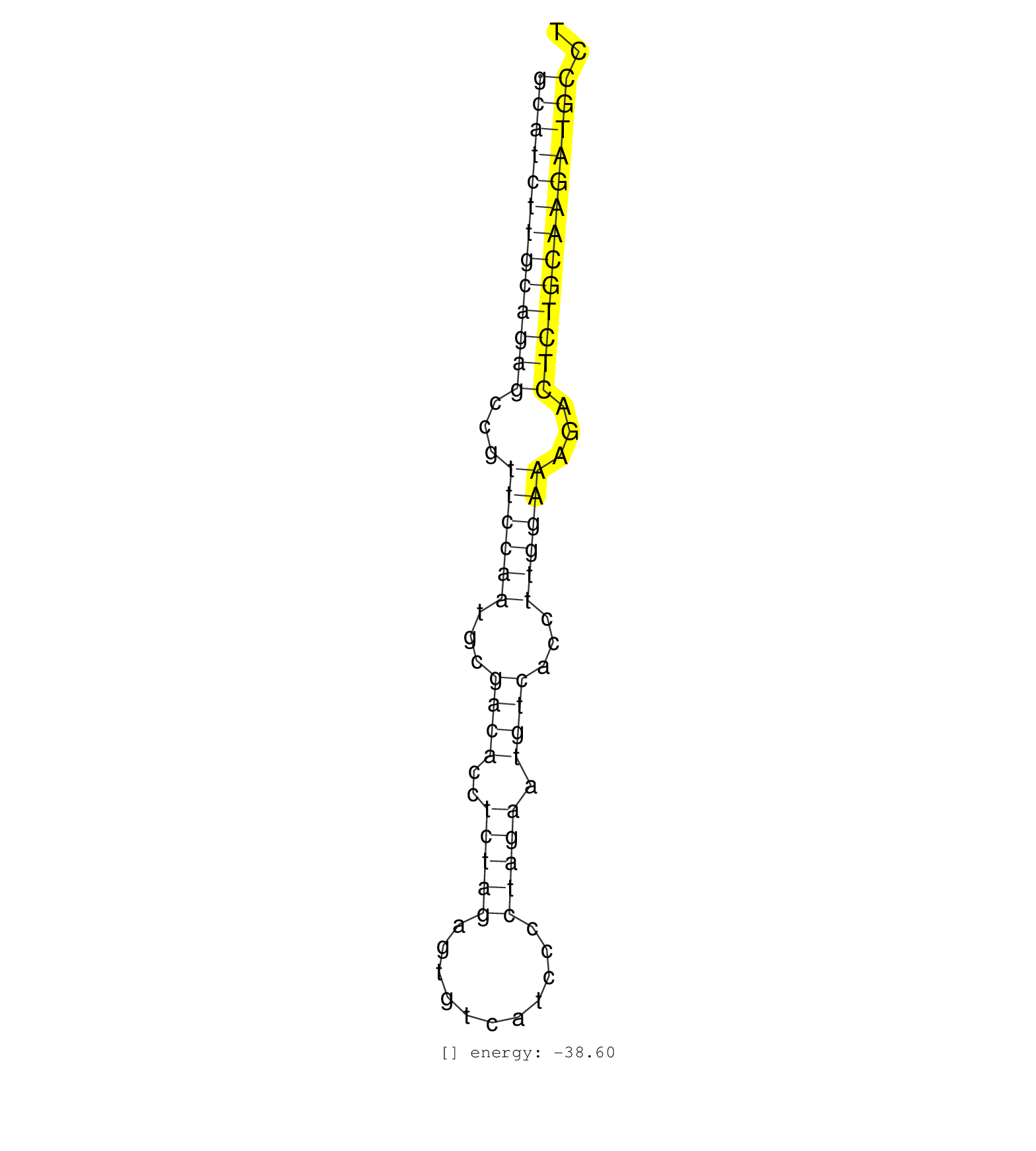
| TGGCCAGAACAGTCTATAACTGAGAATCCCCTCAGACTCTGGGCAGAAAGGCATCTTGCAGAGCCGTTCCAATGCGACACCTCTAGAGTGTCATCCCCTAGAATGTCACCTTGGAAAGACTCTGCAAGATGCCTTGTTAACCATCATGGACCCTGTTAACCATTGTGGGCTAGCATGGTTCTAC ..................................................(((((((((((((...((((((...((((..(((((...........))))).))))...))))))...))))))))))))).................................................... ..................................................51.................................................................................134................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR139188(SRX050640) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR139203(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR189785 | SRR038854(GSM458537) MM653. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139164(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTa................................................ | 22 | A | 31.00 | 12.00 | 27.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCT.................................................. | 20 | 1 | 17.00 | 17.00 | 16.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTt................................................ | 22 | T | 13.00 | 12.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTT................................................. | 21 | 1 | 12.00 | 12.00 | 11.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTaa............................................... | 23 | AA | 11.00 | 12.00 | 8.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCa.................................................. | 20 | A | 7.00 | 3.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTGa............................................... | 23 | A | 5.00 | 4.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTtt............................................... | 23 | TT | 4.00 | 12.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTG................................................ | 22 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCC................................................... | 19 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTGT............................................... | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTata............................................... | 23 | ATA | 2.00 | 17.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCGTTCCAATGCGACACCTCTAG.................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTaa................................................ | 22 | AA | 1.00 | 17.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTta............................................... | 23 | TA | 1.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTatt............................................... | 23 | ATT | 1.00 | 17.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................TAGAATGTCACCTTGGAAAGACa............................................................... | 23 | A | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCTTag............................................... | 23 | AG | 1.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................AAAGACTCTGCAAGATGCCata................................................ | 22 | ATA | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CAGACTCTGGGCAGAA........................................................................................................................................ | 16 | 5 | 0.40 | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | 0.40 | - | - |
| TGGCCAGAACAGTCTATAACTGAGAATCCCCTCAGACTCTGGGCAGAAAGGCATCTTGCAGAGCCGTTCCAATGCGACACCTCTAGAGTGTCATCCCCTAGAATGTCACCTTGGAAAGACTCTGCAAGATGCCTTGTTAACCATCATGGACCCTGTTAACCATTGTGGGCTAGCATGGTTCTAC ..................................................(((((((((((((...((((((...((((..(((((...........))))).))))...))))))...))))))))))))).................................................... ..................................................51.................................................................................134................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR139188(SRX050640) FLASHPage purified small RNA (~15-40nt) from . (lung) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR139203(SRX050636) FLASHPage purified small RNA (~15-40nt) from . (spleen) | SRR189785 | SRR038854(GSM458537) MM653. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR139164(SRX050631) FLASHPage purified small RNA (~15-40nt) from . (heart) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......acCAGTCTATAACTGAG................................................................................................................................................................ | 17 | ac | 3.00 | 0.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................tAAGGCATCTTGCAGAGCCGTT.................................................................................................................... | 22 | t | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| .......................................................................ttctGACACCTCTAGAGTG.............................................................................................. | 19 | ttct | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......ccCAGTCTATAACTGAG................................................................................................................................................................ | 17 | cc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GGCTAGCATGGTTCT.. | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |
| ...............................TCAGACTCTGGGCAG.......................................................................................................................................... | 15 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |